This project performs an exploratory data analysis (EDA) on animal intake and outcome data from the Austin Animal Center. The dataset is sourced from the official City of Austin Open Data Portal.
📦 Importing Necessary Packages
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib as mpl
import matplotlib.pyplot as plt
import missingno as msno
import holoviews as hv
import plotly
import pywaffle as waff
import httpimport
import alluvial
plt.style.use('ggplot')
red = (226/255, 74/255, 51/255)
blue = (52/255, 138/255, 189/255)
📂 Loading Data from csv Files
intakes_df = pd.read_csv('intakes.csv')
outcomes_df = pd.read_csv('outcomes.csv')
📊 Dataset Overview
print('intakes.csv')
display(intakes_df.head(3))
print('outcomes.csv')
display(outcomes_df.head(3))
intakes.csv
| Animal ID | Name | DateTime | MonthYear | Found Location | Intake Type | Intake Condition | Animal Type | Sex upon Intake | Age upon Intake | Breed | Color | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | A786884 | *Brock | 01/03/2019 04:19:00 PM | January 2019 | 2501 Magin Meadow Dr in Austin (TX) | Stray | Normal | Dog | Neutered Male | 2 years | Beagle Mix | Tricolor |
| 1 | A706918 | Belle | 07/05/2015 12:59:00 PM | July 2015 | 9409 Bluegrass Dr in Austin (TX) | Stray | Normal | Dog | Spayed Female | 8 years | English Springer Spaniel | White/Liver |
| 2 | A724273 | Runster | 04/14/2016 06:43:00 PM | April 2016 | 2818 Palomino Trail in Austin (TX) | Stray | Normal | Dog | Intact Male | 11 months | Basenji Mix | Sable/White |
outcomes.csv
| Animal ID | Name | DateTime | MonthYear | Date of Birth | Outcome Type | Outcome Subtype | Animal Type | Sex upon Outcome | Age upon Outcome | Breed | Color | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | A794011 | Chunk | 05/08/2019 06:20:00 PM | May 2019 | 05/02/2017 | Rto-Adopt | NaN | Cat | Neutered Male | 2 years | Domestic Shorthair Mix | Brown Tabby/White |
| 1 | A776359 | Gizmo | 07/18/2018 04:02:00 PM | Jul 2018 | 07/12/2017 | Adoption | NaN | Dog | Neutered Male | 1 year | Chihuahua Shorthair Mix | White/Brown |
| 2 | A821648 | NaN | 08/16/2020 11:38:00 AM | Aug 2020 | 08/16/2019 | Euthanasia | NaN | Other | Unknown | 1 year | Raccoon | Gray |
print('intakes.csv:')
intakes_df.info()
print('\noutcomes.csv:')
outcomes_df.info()
intakes.csv:
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 138585 entries, 0 to 138584
Data columns (total 12 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Animal ID 138585 non-null object
1 Name 97316 non-null object
2 DateTime 138585 non-null object
3 MonthYear 138585 non-null object
4 Found Location 138585 non-null object
5 Intake Type 138585 non-null object
6 Intake Condition 138585 non-null object
7 Animal Type 138585 non-null object
8 Sex upon Intake 138584 non-null object
9 Age upon Intake 138585 non-null object
10 Breed 138585 non-null object
11 Color 138585 non-null object
dtypes: object(12)
memory usage: 12.7+ MB
outcomes.csv:
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 138769 entries, 0 to 138768
Data columns (total 12 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Animal ID 138769 non-null object
1 Name 97514 non-null object
2 DateTime 138769 non-null object
3 MonthYear 138769 non-null object
4 Date of Birth 138769 non-null object
5 Outcome Type 138746 non-null object
6 Outcome Subtype 63435 non-null object
7 Animal Type 138769 non-null object
8 Sex upon Outcome 138768 non-null object
9 Age upon Outcome 138764 non-null object
10 Breed 138769 non-null object
11 Color 138769 non-null object
dtypes: object(12)
memory usage: 12.7+ MB
Data Size
The intakes dataset initially contains 138,585 records, and outcomes has 138,769 records.
🔎 Feature List
Both datasets have 12 features. Pandas identifies all of them as object (string) type.
The datasets share the following common features (data types characteristic of the values are in brackets):
- Animal ID [str] - Unique animal identifier
- Name [str] - Animal’s name
- DateTime [datetime] - Timestamp of intake (in) or outcome (out)
- MonthYear [date] - Month and year of intake (in) or outcome (out)
- Animal Type [categorical nominal] - Species of animal
- Intake-Outcome Type [categorical nominal] - Method of intake (in) or outcome (out)
- Sex upon Intake-Outcome [categorical nominal] - Animal’s sex (including spay/neuter status)
- Age upon Intake-Outcome [int] - Animal’s age at intake (in) or outcome (out)
- Breed [categorical nominal] - Animal’s breed
- Color [categorical nominal] - Animal’s color
The intakes dataset also includes:
- Found Location [categorical nominal] - Location where the animal was found
- Intake Condition [categorical nominal] - Animal’s condition upon intake
The outcomes dataset also includes:
- Date of Birth [date] - Animal’s date of birth
- Outcome Subtype [categorical nominal] - Subcategory of outcome type
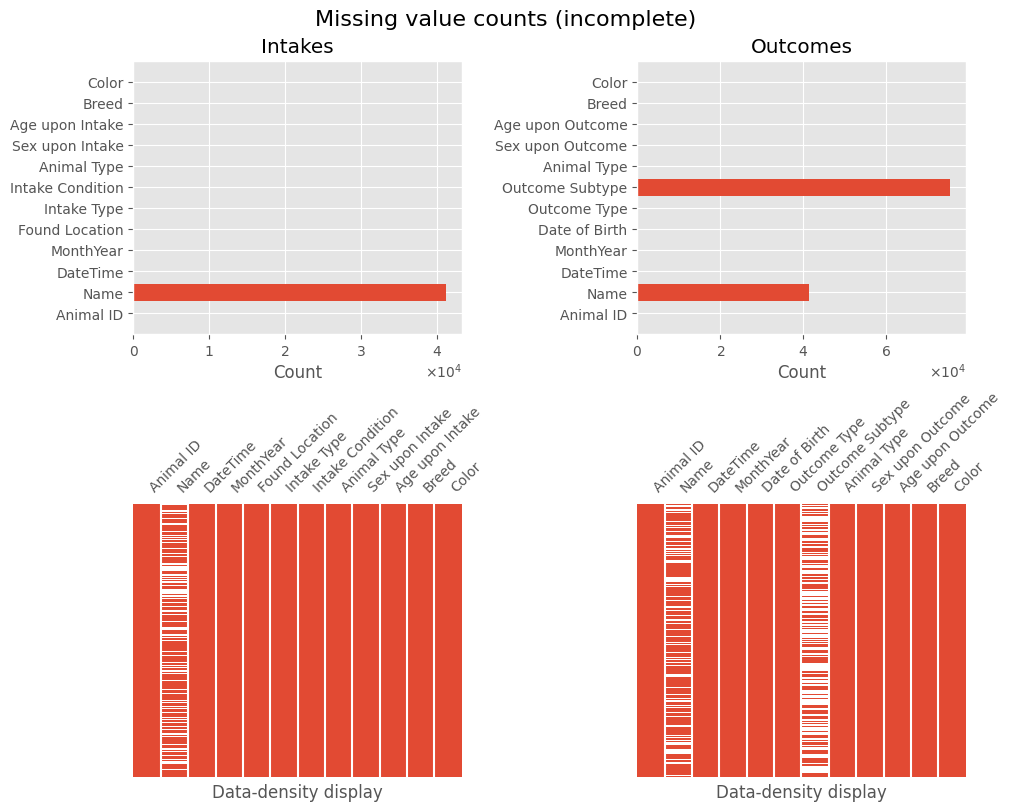
❓ Missing Values
The initial data information shows missing values in:
intakes:Name,Sex upon Intakeoutcomes:Name,Outcome Type,Outcome Subtype,Sex upon Outcome,Age upon Outcome
More missing values will be addressed during individual feature processing.
def subplot_missing_values(ax, data, title):
ax.barh(data.index, data)
ax.set_title(title)
ax.set_xlabel('Count')
ax.ticklabel_format(axis='x', style='sci', scilimits=(0,0), useMathText=True)
def subplot_missing_values_matrix(ax, data):
msno.matrix(data, fontsize=10, sparkline=False, ax=ax, color=red)
ax.set_xlabel('Data-density display')
ax.get_yaxis().set_visible(False)
def plot_missing(df1, name1, df2, name2, *, title='Missing value counts (incomplete)'):
fig, axes = plt.subplots(2, 2, figsize=(10, 8), layout='constrained')
fig.suptitle(title, fontsize=16)
subplot_missing_values(axes[0][0], df1.isna().sum(axis=0), name1)
subplot_missing_values(axes[0][1], df2.isna().sum(axis=0), name2)
subplot_missing_values_matrix(axes[1][0], df1)
subplot_missing_values_matrix(axes[1][1], df2)
plot_missing(intakes_df, 'Intakes', outcomes_df, 'Outcomes')
Unique Feature Values
print('intakes.csv:')
display(intakes_df.nunique())
print(intakes_df.apply(lambda col: col.unique()))
print('\noutcomes.csv:')
display(outcomes_df.nunique())
print(outcomes_df.apply(lambda col: col.unique()))
intakes.csv:
Animal ID 123890
Name 23544
DateTime 97442
MonthYear 103
Found Location 58367
Intake Type 6
Intake Condition 15
Animal Type 5
Sex upon Intake 5
Age upon Intake 54
Breed 2741
Color 616
dtype: int64
Animal ID [A786884, A706918, A724273, A665644, A682524, ...
Name [*Brock, Belle, Runster, nan, Rio, Odin, Beowu...
DateTime [01/03/2019 04:19:00 PM, 07/05/2015 12:59:00 P...
MonthYear [January 2019, July 2015, April 2016, October ...
Found Location [2501 Magin Meadow Dr in Austin (TX), 9409 Blu...
Intake Type [Stray, Owner Surrender, Public Assist, Wildli...
Intake Condition [Normal, Sick, Injured, Pregnant, Nursing, Age...
Animal Type [Dog, Cat, Other, Bird, Livestock]
Sex upon Intake [Neutered Male, Spayed Female, Intact Male, In...
Age upon Intake [2 years, 8 years, 11 months, 4 weeks, 4 years...
Breed [Beagle Mix, English Springer Spaniel, Basenji...
Color [Tricolor, White/Liver, Sable/White, Calico, T...
dtype: object
outcomes.csv:
Animal ID 124068
Name 23425
DateTime 115364
MonthYear 103
Date of Birth 7576
Outcome Type 9
Outcome Subtype 26
Animal Type 5
Sex upon Outcome 5
Age upon Outcome 54
Breed 2749
Color 619
dtype: int64
Animal ID [A794011, A776359, A821648, A720371, A674754, ...
Name [Chunk, Gizmo, nan, Moose, Princess, Quentin, ...
DateTime [05/08/2019 06:20:00 PM, 07/18/2018 04:02:00 P...
MonthYear [May 2019, Jul 2018, Aug 2020, Feb 2016, Mar 2...
Date of Birth [05/02/2017, 07/12/2017, 08/16/2019, 10/08/201...
Outcome Type [Rto-Adopt, Adoption, Euthanasia, Transfer, Re...
Outcome Subtype [nan, Partner, Foster, SCRP, Out State, Suffer...
Animal Type [Cat, Dog, Other, Bird, Livestock]
Sex upon Outcome [Neutered Male, Unknown, Intact Male, Spayed F...
Age upon Outcome [2 years, 1 year, 4 months, 6 days, 7 years, 2...
Breed [Domestic Shorthair Mix, Chihuahua Shorthair M...
Color [Brown Tabby/White, White/Brown, Gray, Buff, O...
dtype: object
The number of unique Animal IDs does not match the number of rows in their respective datasets, indicating some animals return to the shelter. Also, Age upon Intake/Outcome has inconsistent units (years, months, days), which needs standardization.
🧹 Data Exploration and Cleaning
print(pd.concat([intakes_df.columns.to_series(), outcomes_df.columns.to_series()]).drop_duplicates().tolist())
['Animal ID', 'Name', 'DateTime', 'MonthYear', 'Found Location', 'Intake Type', 'Intake Condition', 'Animal Type', 'Sex upon Intake', 'Age upon Intake', 'Breed', 'Color', 'Date of Birth', 'Outcome Type', 'Outcome Subtype', 'Sex upon Outcome', 'Age upon Outcome']
The cleaning process involves:
- Manually inspecting features for various representations of missing values, then replacing them with
NaN. - Transforming data for consistency (e.g., converting age to a common unit, standardizing date formats).
- Converting columns to appropriate data types (e.g., category, int, datetime).
- Removing redundant features and splitting complex ones.
First, we’ll remove duplicate records.
intakes_df = intakes_df.drop_duplicates()
outcomes_df = outcomes_df.drop_duplicates()
print(intakes_df.shape[0], outcomes_df.shape[0])
138565 138752
After dropping duplicates, intakes_df has 138,565 records and outcomes_df has 138,752 records.
Animal ID
print(sum(intakes_df['Animal ID'].str.startswith('A') == False),
sum(outcomes_df['Animal ID'].str.startswith('A') == False))
0 0
All Animal ID values start with “A”. The IDs are consistent and have no missing values.
Name
print(intakes_df['Name'].iloc[:10].tolist())
['*Brock', 'Belle', 'Runster', nan, 'Rio', 'Odin', 'Beowulf', '*Ella', 'Mumble', nan]
Some names begin with an asterisk. We’ll remove this character for consistency. Also, some entries in the Name column duplicate the Animal ID; these will be replaced with NaN.
intakes_df['Name'] = intakes_df.loc[:, 'Name'].str.removeprefix('*')
outcomes_df['Name'] = intakes_df.loc[:, 'Name'].str.removeprefix('*')
mask = intakes_df['Name'] == intakes_df['Animal ID']
intakes_df.loc[mask, 'Name'] = np.nan
DateTime, Date of Birth
The DateTime and Date of Birth features contain time values, so we convert them to the datetime64 data type.
intakes_df['DateTime'] = pd.to_datetime(intakes_df.loc[:, 'DateTime'], format='%m/%d/%Y %H:%M:%S %p')
outcomes_df['DateTime'] = pd.to_datetime(outcomes_df.loc[:, 'DateTime'], format='%m/%d/%Y %H:%M:%S %p')
outcomes_df['Date of Birth'] = pd.to_datetime(outcomes_df.loc[:, 'Date of Birth'], format='%m/%d/%Y')
MonthYear
# porovnání hodnot v MonthYear a DateTime
print(sum(intakes_df['MonthYear'].str.split(expand=True)[0] != intakes_df['DateTime'].dt.month_name()),
sum(intakes_df['MonthYear'].str.split(expand=True)[1].astype('int32') != intakes_df['DateTime'].dt.year))
0 0
The MonthYear feature contains redundant information already present in DateTime. We can drop this column.
intakes_df = intakes_df.drop(columns=['MonthYear'], errors='ignore')
outcomes_df = outcomes_df.drop(columns=['MonthYear'], errors='ignore')
Intake-Outcome Type, Intake Condition, Animal Type, Breed, Color, Outcome Subtype, Found Location
print('Animal Type:\t\t', *intakes_df['Animal Type'].unique())
print('Intake Type:\t\t', *intakes_df['Intake Type'].unique())
print('Outcome Type:\t\t', *outcomes_df['Outcome Type'].unique())
print('Intake Condition:\t', *intakes_df['Intake Condition'].unique())
print('Outcome Subtype:\t', *outcomes_df['Outcome Subtype'].unique()[:10], '...')
print('Breed:\t\t\t', *intakes_df['Breed'].unique()[:5], '...')
print('Color:\t\t\t', *intakes_df['Color'].unique()[:10], '...')
print('Found Location:\t\t', *intakes_df['Found Location'].unique()[:4], '...')
Animal Type: Dog Cat Other Bird Livestock
Intake Type: Stray Owner Surrender Public Assist Wildlife Euthanasia Request Abandoned
Outcome Type: Rto-Adopt Adoption Euthanasia Transfer Return to Owner Died Disposal Missing Relocate nan
Intake Condition: Normal Sick Injured Pregnant Nursing Aged Medical Other Neonatal Feral Behavior Med Urgent Space Med Attn Panleuk
Outcome Subtype: nan Partner Foster SCRP Out State Suffering Underage Snr Rabies Risk In Kennel ...
Breed: Beagle Mix English Springer Spaniel Basenji Mix Domestic Shorthair Mix Doberman Pinsch/Australian Cattle Dog ...
Color: Tricolor White/Liver Sable/White Calico Tan/Gray Chocolate Black Brown Tabby Black/White Cream Tabby ...
Found Location: 2501 Magin Meadow Dr in Austin (TX) 9409 Bluegrass Dr in Austin (TX) 2818 Palomino Trail in Austin (TX) Austin (TX) ...
Values in these features appear to be in order. We will convert these features to categorical type. We also observed various ways missing values are represented ('', nan, Unknown); these will all be replaced with NaN.
for nanstr in ['', 'nan', 'Unknown']:
intakes_df = intakes_df.replace(nanstr, np.nan)
outcomes_df = outcomes_df.replace(nanstr, np.nan)
in_cols = ['Intake Type', 'Intake Condition', 'Animal Type', 'Breed', 'Color', 'Found Location']
out_cols = ['Outcome Type', 'Outcome Subtype', 'Animal Type', 'Breed', 'Color']
intakes_df[in_cols] = intakes_df[in_cols].astype('category')
outcomes_df[out_cols] = outcomes_df[out_cols].astype('category')
def subplot_cat_values(ax, series, *, rot=False, ylabel=True):
counts = series.value_counts()
ax.bar(counts.index, counts)
ax.set_xticks(counts.index)
ax.tick_params(labelsize=9)
if rot:
ax.tick_params(rotation=45)
ax.set_xlabel(series.name, fontsize=12)
if ylabel:
ax.set_ylabel('Count', fontsize=12)
ax.ticklabel_format(axis='y', style='sci', scilimits=(0,0), useMathText=True)
def plot_cat_values(df, title, c1, c2, c3, rot=False):
fig = plt.figure( figsize=(10, 8), layout='constrained')
gs = fig.add_gridspec(2, 3)
ax1 = fig.add_subplot(gs[0, 0])
ax2 = fig.add_subplot(gs[0, 1:3])
ax3 = fig.add_subplot(gs[1, :])
fig.suptitle(title, fontsize=18)
subplot_cat_values(ax1, df[c1])
subplot_cat_values(ax2, df[c2], rot=rot)
subplot_cat_values(ax3, df[c3], rot=rot)
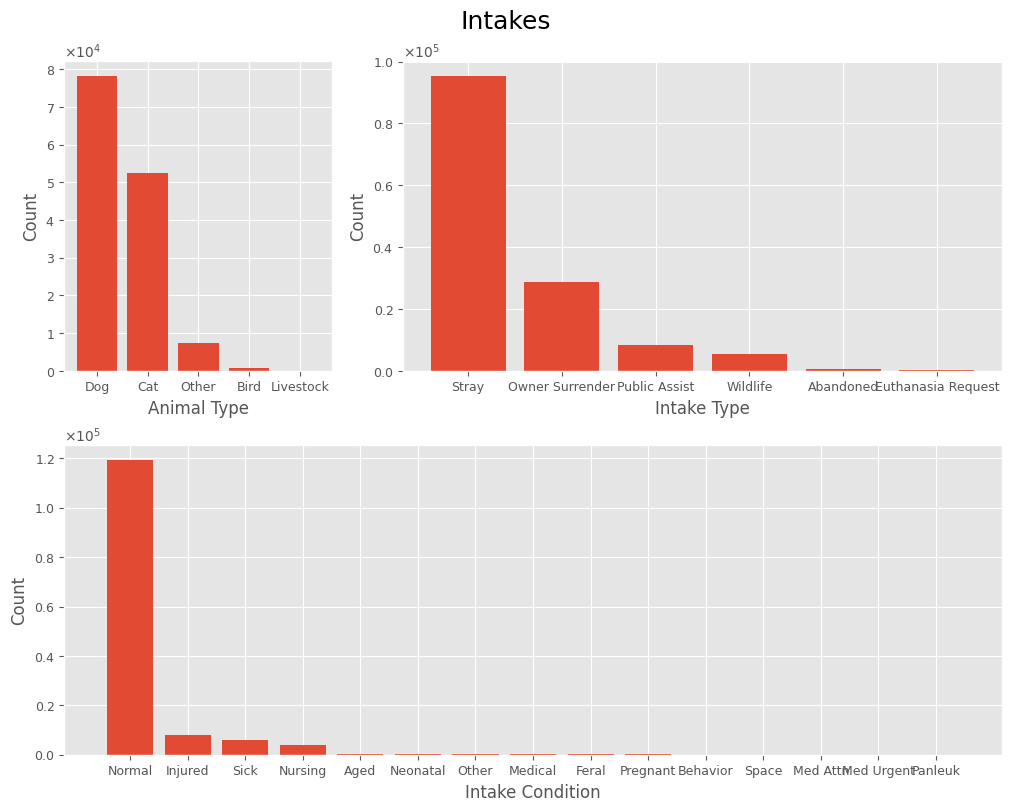
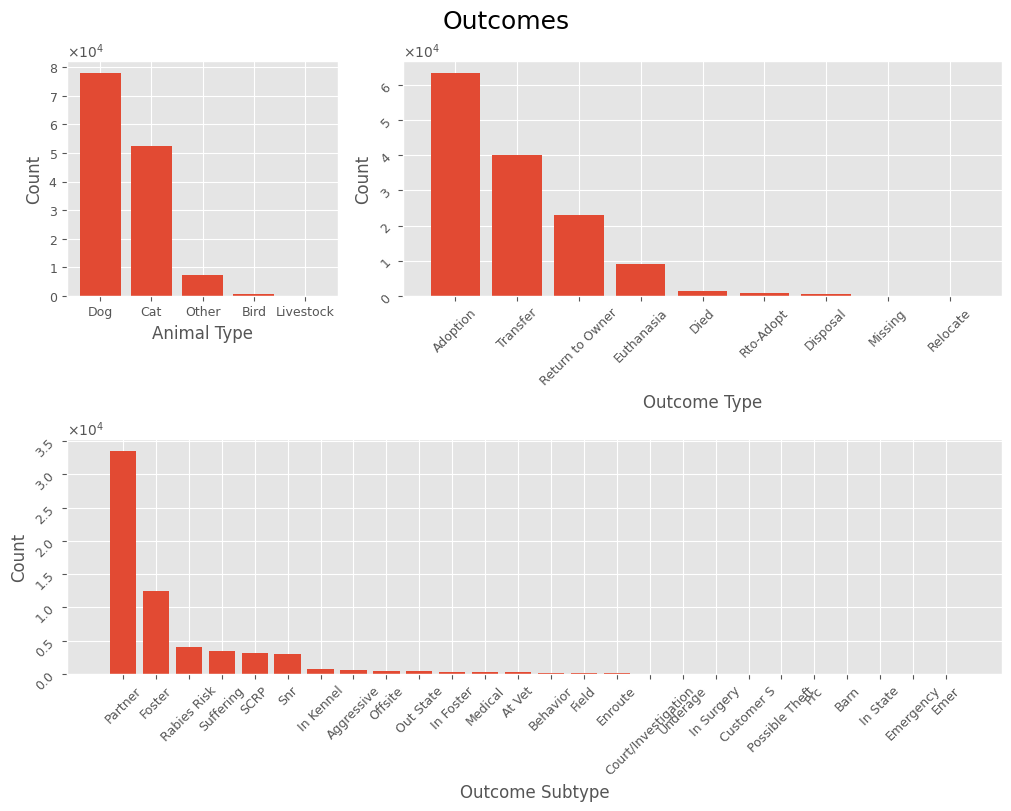
For features with fewer categories, we can visualize the frequency of each value to gain a better understanding of the data. No conclusions are drawn at this stage.
plot_cat_values(intakes_df, 'Intakes', *['Animal Type', 'Intake Type', 'Intake Condition'], rot=False)
plot_cat_values(outcomes_df, 'Outcomes', *['Animal Type', 'Outcome Type', 'Outcome Subtype'], rot=True)
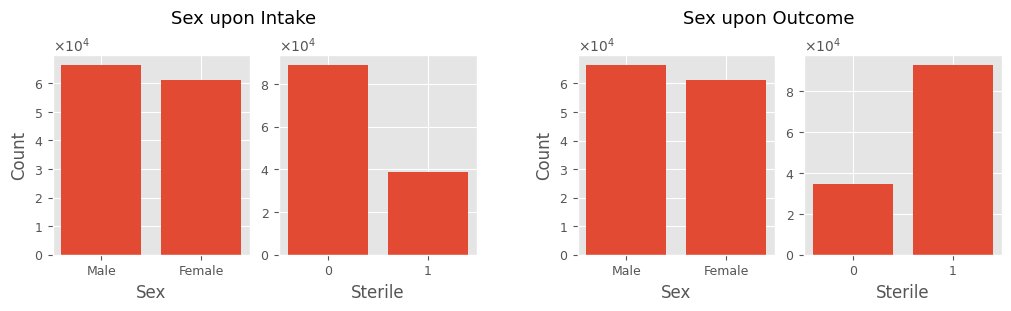
Sex upon Intake-Outcome
The Sex upon Intake and Sex upon Outcome features can be split into two: Sex (categorical) and Sterile (boolean). This will make them easier to work with.
We’ll perform the following transformations to create the new Sterile feature:
Neutered Male,Spayed Female->Sterile:TrueIntact Male,Intact Female->Sterile:False
The sex will be separated and assigned to a new Sex column. The original feature will then be dropped. Finally, both new features will be set as categorical types.
Any data inconsistencies will be discussed in more detail in the final analysis section.
print('Sex upon Intake:\t', intakes_df['Sex upon Intake'].unique())
print('Sex upon Outcome:\t', outcomes_df['Sex upon Outcome'].unique())
Sex upon Intake: ['Neutered Male' 'Spayed Female' 'Intact Male' 'Intact Female' nan]
Sex upon Outcome: ['Neutered Male' nan 'Intact Male' 'Spayed Female' 'Intact Female']
# split
intakes_df[['Sterile', 'Sex']] = intakes_df['Sex upon Intake'].str.split(expand=True)
outcomes_df[['Sterile', 'Sex']] = outcomes_df['Sex upon Outcome'].str.split(expand=True)
sterilMap = {'Neutered': True, 'Spayed': True, 'Intact': False}
intakes_df['Sterile'] = intakes_df['Sterile'].map(sterilMap)
outcomes_df['Sterile'] = outcomes_df['Sterile'].map(sterilMap)
# drop
intakes_df = intakes_df.drop(columns=['Sex upon Intake'])
outcomes_df = outcomes_df.drop(columns=['Sex upon Outcome'])
cols = ['Sterile', 'Sex']
intakes_df[cols] = intakes_df[cols].astype('category')
outcomes_df[cols] = outcomes_df[cols].astype('category')
fig = plt.figure(figsize=(10, 3), layout='constrained')
(fig1, fig2) = fig.subfigures(1, 2, wspace=0.1)
axes = fig1.subplots(1, 2)
fig1.suptitle('Sex upon Intake', fontsize=13)
subplot_cat_values(axes[0], intakes_df['Sex'])
subplot_cat_values(axes[1], intakes_df['Sterile'], ylabel=False)
axes = fig2.subplots(1, 2)
fig2.suptitle('Sex upon Outcome', fontsize=13)
subplot_cat_values(axes[0], outcomes_df['Sex'])
subplot_cat_values(axes[1], outcomes_df['Sterile'], ylabel=False)
Age upon Intake-Outcome
intakes_df['Age upon Intake'].unique()
array(['2 years', '8 years', '11 months', '4 weeks', '4 years', '6 years',
'6 months', '5 months', '14 years', '1 month', '2 months',
'18 years', '9 years', '4 months', '1 year', '3 years', '4 days',
'1 day', '5 years', '2 weeks', '15 years', '7 years', '3 weeks',
'3 months', '12 years', '1 week', '9 months', '10 years',
'10 months', '7 months', '8 months', '1 weeks', '5 days',
'0 years', '2 days', '11 years', '17 years', '3 days', '13 years',
'5 weeks', '19 years', '6 days', '16 years', '20 years',
'-1 years', '22 years', '23 years', '-2 years', '21 years',
'-3 years', '25 years', '24 years', '30 years', '28 years'],
dtype=object)
The unique values show negative age entries, which are errors. These records will be corrected, and all age values will be converted to days for consistency.
The process is:
- Separate the numerical value from the unit.
- Convert to a non-negative number.
- Transform the age to total days based on its unit.
During this process, the age features will be converted to a numerical type. Animals listed as 0 years will be interpreted as newborns.
tmp_in = intakes_df['Age upon Intake'].str.split(expand=True)
tmp_out = outcomes_df['Age upon Outcome'].str.split(expand=True)
tmp_in[0] = tmp_in[0].astype('Int64').abs() # nullable integer
tmp_out[0] = tmp_out[0].astype('Int64').abs()
print(*tmp_in[1].unique())
print(*tmp_out[1].unique())
years months weeks month year days day week
years year months days weeks month day week nan
def convert_ages(tmp_df):
yearMask = tmp_df[1].isin(['years', 'year'])
monthMask = tmp_df[1].isin(['months', 'month'])
weekMask = tmp_df[1].isin(['weeks', 'week'])
tmp_df.loc[yearMask, 0] = tmp_df.loc[yearMask, 0] * 365
tmp_df.loc[monthMask, 0] = tmp_df.loc[monthMask, 0] * 30
tmp_df.loc[weekMask, 0] = tmp_df.loc[weekMask, 0] * 7
return tmp_df[0]
intakes_df['Age upon Intake'] = convert_ages(tmp_in)
outcomes_df['Age upon Outcome'] = convert_ages(tmp_out)
def subplot_age(ax, data, title):
ax.set_title(title)
ax.hist(data, bins=15)
ax.set_ylabel('Count')
ax.set_xlabel('Age [years]')
ax.ticklabel_format(axis='y', style='sci', scilimits=(0,0), useMathText=True)
fig, axes = plt.subplots(1, 2, figsize=(10, 3), layout='constrained')
subplot_age(axes[0], intakes_df['Age upon Intake'].dropna()/365, 'Age upon Intake')
subplot_age(axes[1], outcomes_df['Age upon Outcome'].dropna()/365, 'Age upon Outcome')
💡 Summary
We have converted the features of both datasets to their appropriate data types:
Animal IDandNameremain asstring(object).DateTimeandDate of Birthare converted todatetime(datetime64).- Age features are transformed to numerical
int64. - All other features are converted to
categorytype. - The redundant
MonthYearfeature was removed. - The
Sex upon Intake-Outcomefeature was split into two meaningful features:SexandSterile. - Erroneous data (negative ages, typos) were corrected, and missing values were handled.
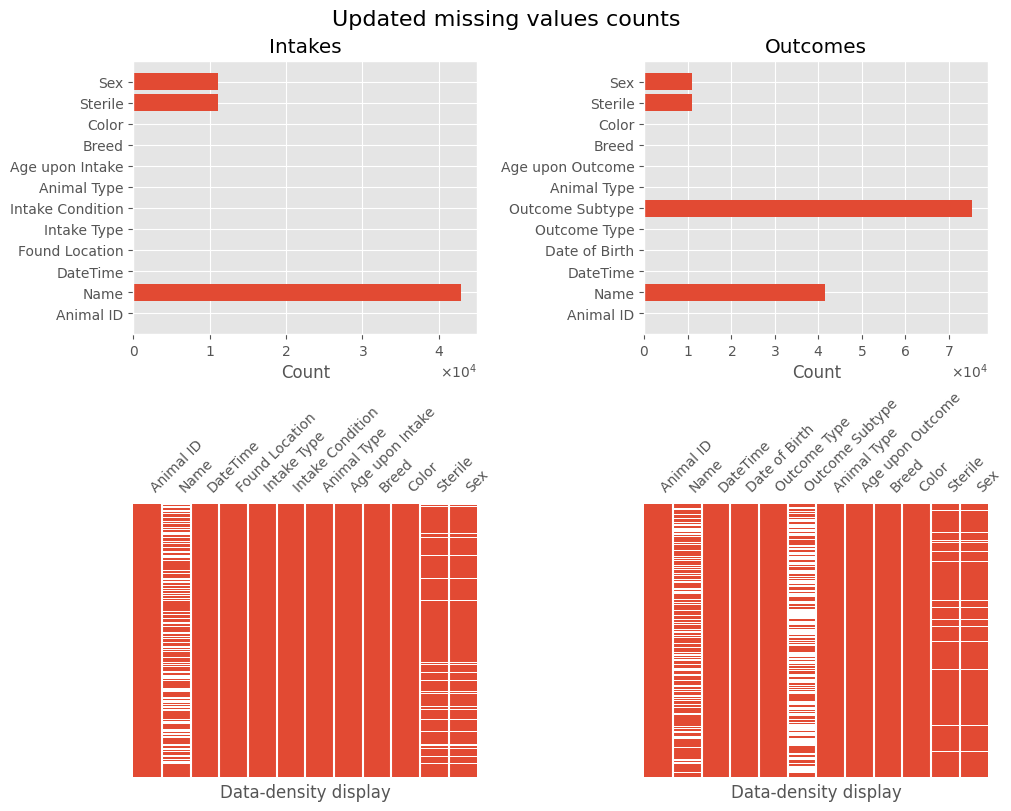
We now have a much clearer understanding of the missing data. The pre-processed dataframes are intakes_df and outcomes_df.
plot_missing(intakes_df, 'Intakes', outcomes_df, 'Outcomes', title='Updated missing values counts')
🖊️ Descriptive Statistics
def print_univar_num_stats(series):
print(series.name, 'statistics')
print()
print('min:\t', series.min())
print('max:\t', series.max())
print('mean:\t', series.mean())
print('median:\t', series.median())
print('range:\t', series.max() - series.min())
print()
print('lower quartile:\t', series.quantile(0.25))
print('upper quartile:\t', series.quantile(0.75))
print('IQR:\t\t', series.quantile(0.75) - series.quantile(0.25))
print()
print('variance:\t', series.var())
print('std. variation:\t', series.std())
print('skewness:\t', series.skew())
print('kurtosis:\t', series.kurtosis())
def print_univar_dt_stats(series):
print(series.name, 'statistics')
print()
print('min:\t', series.min())
print('max:\t', series.max())
print('mode:\t', list(series.mode()))
print('range:\t', series.max() - series.min())
def plot_univar_num(title1, title2, xlabel, ylabel, data, *, bins=15, xlim=None, scinot=False, showfliers=True):
fig, axes = plt.subplots(1, 2, figsize=(10, 4), layout='constrained', gridspec_kw={'wspace': 0.1})
ax = axes[0]
ax.set_title(title1)
ax.set_ylabel(ylabel)
ax.set_xlabel(xlabel)
if scinot:
ax.ticklabel_format(axis='y', style='sci', scilimits=(0, 0), useMathText=True)
ax.hist(data, bins=bins)
medianprops = dict(linewidth=2.5, color=(1, 0.38, 0.27))
flierprops = dict(marker='d', markerfacecolor=(1, 0.38, 0.27), markersize=8, markeredgecolor='none')
ax = axes[1]
ax.set_title(title2)
ax.set_xlabel(xlabel)
if xlim != None:
ax.set_xlim(xlim)
ax.boxplot(data, vert=False, widths=[0.4], showfliers=showfliers, flierprops=flierprops, medianprops=medianprops)
ax.get_yaxis().set_visible(False)
🎂 Age upon Intake Feature
This section describes the Age upon Intake and DateTime features (originally from the intakes dataset) using univariate descriptive statistics.
age = intakes_df['Age upon Intake'].dropna()
ageY = intakes_df['Age upon Intake'] / 365
age.info()
<class 'pandas.core.series.Series'>
Index: 138565 entries, 0 to 138584
Series name: Age upon Intake
Non-Null Count Dtype
-------------- -----
138565 non-null Int64
dtypes: Int64(1)
memory usage: 2.2 MB
print_univar_num_stats(ageY)
Age upon Intake statistics
min: 0.0
max: 30.0
mean: 2.0278033206313832
median: 1.0
range: 30.0
lower quartile: 0.1643835616438356
upper quartile: 2.0
IQR: 1.8356164383561644
variance: 8.170721564564788
std. variation: 2.8584474045475785
skewness: 2.3340016328003066
kurtosis: 5.968203952133479
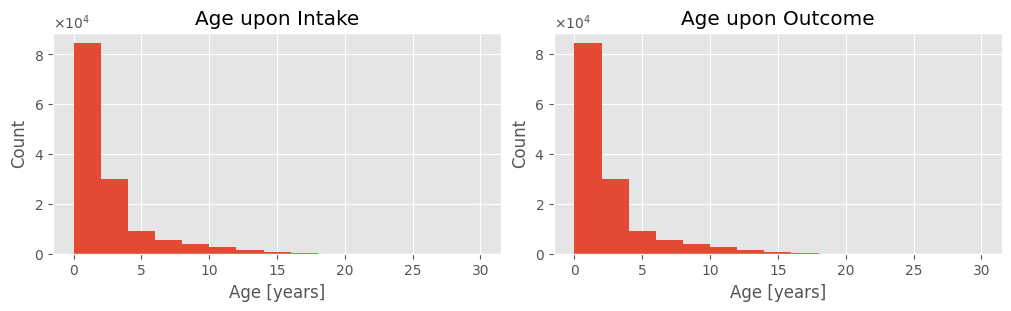
The Age upon Intake feature represents the animal’s age at intake in days, after preprocessing. Records with missing values were excluded from this analysis.
- Basic descriptive statistics show animal ages at intake range from 0 (newborns) to 30 years.
- The mean age is slightly over 2 years, while the median is 1 year.
- Despite a total range of 30 years, the interquartile range (IQR) is only 1.8 years, indicating that the majority of animals are young.
- A positive skewness coefficient indicates a right-skewed distribution.
- The high skewness coefficient suggests most values are clustered near the mean of 2 years.
The following histogram visually confirms these observations. The cropped boxplot clearly shows that despite an average age of 2, most (75%) animals are younger. The mean is skewed by outliers, making the median a better representation of the typical age of animals at the shelter.
plot_univar_num('Age upon Intake', 'Age upon Intake < 6 (focused)', 'Age [years]', 'Count', ageY, xlim=(-0.5, 6.5), scinot=True)
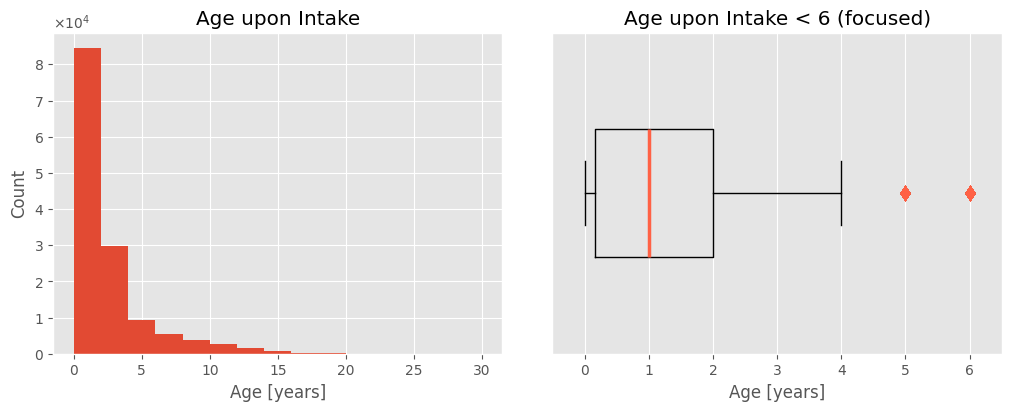
intakes_df[ageY == 30]
| Animal ID | Name | DateTime | Found Location | Intake Type | Intake Condition | Animal Type | Age upon Intake | Breed | Color | Sterile | Sex | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 132371 | A842878 | Sunshine | 2021-10-30 10:07:00 | 3008 West Avenue in Austin (TX) | Owner Surrender | Other | Bird | 10950 | Macaw | Blue/Gold | False | Female |
The oldest animal recorded in the shelter is a 30-year-old female Macaw named Sunshine.
📅 Datetime Feature (intakes)
This section describes the DateTime feature from the intakes dataset using univariate descriptive statistics.
def dt_to_counts(dts):
dtc = pd.to_datetime(dts).dt.date.value_counts()
# fill days with no intakes
idx = pd.date_range(dtc.index.min(), dtc.index.max())
dtc.index = pd.DatetimeIndex(dtc.index)
dtc = dtc.reindex(idx, fill_value=0)
return dtc
dt = intakes_df['DateTime']
dtcount = dt_to_counts(dt)
dt.info()
<class 'pandas.core.series.Series'>
Index: 138565 entries, 0 to 138584
Series name: DateTime
Non-Null Count Dtype
-------------- -----
138565 non-null datetime64[ns]
dtypes: datetime64[ns](1)
memory usage: 6.1 MB
print_univar_dt_stats(dt)
DateTime statistics
min: 2013-10-01 01:12:00
max: 2022-04-27 07:54:00
mode: [Timestamp('2014-07-09 12:58:00'), Timestamp('2016-09-23 12:00:00')]
range: 3130 days 06:42:00
print_univar_num_stats(dtcount)
count statistics
min: 0
max: 140
mean: 44.25582880868732
median: 44.0
range: 140
lower quartile: 34.0
upper quartile: 55.0
IQR: 21.0
variance: 331.17574476812825
std. variation: 18.198234660761145
skewness: 0.26627105755009095
kurtosis: 0.7186242354271446
dtcount[dtcount == dtcount.max()]
2014-07-09 140
Freq: D, Name: count, dtype: int64
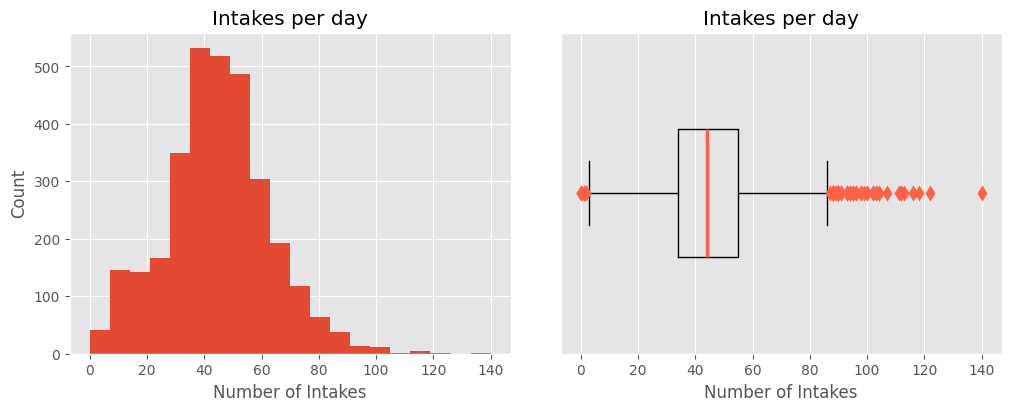
The DateTime feature from the intakes dataset is a temporal feature.
- The earliest recorded animal intake was on October 1, 2013, and the latest was on April 27, 2022.
- The highest number of animal intakes in a single day occurred on July 9, 2014, with 140 new animals added to the system.
- On average, 44 animals are admitted each day, with 50% of days seeing between 34 and 55 animal intakes.
plot_univar_num('Intakes per day', 'Intakes per day', 'Number of Intakes', 'Count', dtcount, bins=20, showfliers=True)
🐾 Animal Type Feature (intakes)
1. Next, we’ll examine three more features using appropriate univariate descriptive statistics.
def plot_univar_cat(title1, title2, xlabel, icons, data, *, prop=1000):
fig, axes = plt.subplots(1, 2, figsize=(10, 4), layout='constrained', gridspec_kw={'wspace': 0.2})
ax = axes[0]
ax.set_title(title1)
ax.set_xlabel(xlabel)
ax.ticklabel_format(axis='x', style='sci', scilimits=(0, 0), useMathText=True)
ax.barh(data.index, data)
ax.invert_yaxis()
val = data / prop
val_freq = val / val.sum()
waff.Waffle.make_waffle(
ax=axes[1],
rows=12,
values=val,
title={'label': title2, 'loc': 'center'},
labels=[f"{k} ({v*100:.2f}%)" for k, v in val_freq.items()],
legend={'bbox_to_anchor': (1.7, 1), 'ncol': 1, 'framealpha': 0},
icons=icons,
font_size=16,
# icon_style='solid',
icon_legend=True,
starting_location='NW',
vertical=True,
cmap_name="Set2"
)
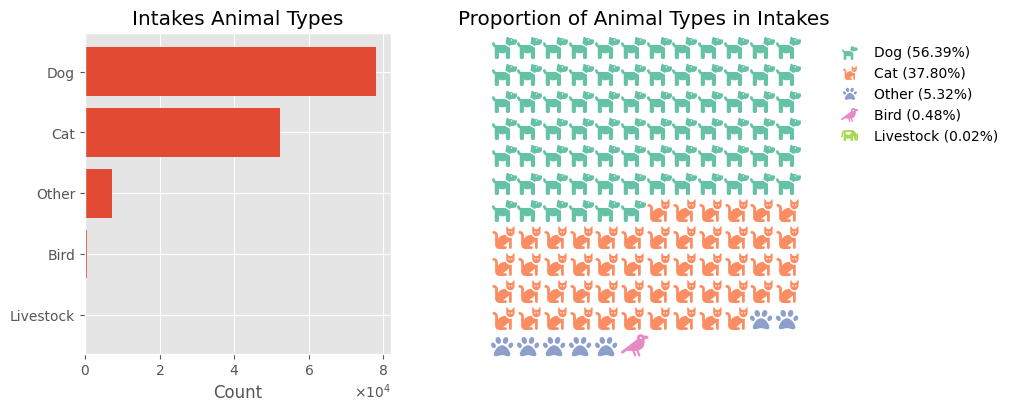
in_type = intakes_df['Animal Type']
in_type_c = in_type.value_counts()
in_type_cnorm = in_type.value_counts(normalize=True)
print('Intakes Animal Type')
display(in_type.info())
Intakes Animal Type
<class 'pandas.core.series.Series'>
Index: 138565 entries, 0 to 138584
Series name: Animal Type
Non-Null Count Dtype
-------------- -----
138565 non-null category
dtypes: category(1)
memory usage: 5.2 MB
None
in_type.describe()
count 138565
unique 5
top Dog
freq 78135
Name: Animal Type, dtype: object
in_type_c
Animal Type
Dog 78135
Cat 52373
Other 7372
Bird 661
Livestock 24
Name: count, dtype: int64
The Animal Type feature is a categorical feature with 5 categories: Bird, Cat, Dog, Livestock, and Other.
- The most common animal type at the shelter is dogs (56%).
- Cats follow, accounting for 37% of intakes.
- Less common animal types include birds, livestock, and “others.”
- Together, dogs and cats comprise over 94% of all recorded intakes.
plot_univar_cat('Intakes Animal Types', 'Proportion of Animal Types in Intakes', 'Count', ['dog', 'cat', 'paw', 'crow', 'cow'], in_type_c)
🌈 Color Feature (intakes)
2. We’ll now examine the Color feature using appropriate univariate descriptive statistics.
intakes_df['Color'].str.split('/', n=-1).value_counts()
Color
[Black, White] 14469
[Black] 11613
[Brown Tabby] 7948
[Brown] 5935
[White] 4866
...
[Black Tabby, Gray] 1
[Yellow, Red] 1
[Seal Point, Cream] 1
[White, Lilac Point] 1
[Brown Tabby, Tan] 1
Name: count, Length: 616, dtype: int64
len(intakes_df['Color'].unique())
616
cols = intakes_df['Color'].str.split('/', n=-1)
cols = pd.Series(np.concatenate(cols.to_numpy().flatten()))
cols = cols.value_counts()
cols.index, len(cols.index)
(Index(['White', 'Black', 'Brown', 'Tan', 'Brown Tabby', 'Blue', 'Orange Tabby',
'Tricolor', 'Brown Brindle', 'Red', 'Gray', 'Blue Tabby', 'Tortie',
'Calico', 'Chocolate', 'Torbie', 'Cream', 'Cream Tabby', 'Fawn',
'Sable', 'Yellow', 'Buff', 'Lynx Point', 'Blue Merle', 'Gray Tabby',
'Seal Point', 'Orange', 'Black Brindle', 'Flame Point', 'Black Tabby',
'Blue Tick', 'Brown Merle', 'Gold', 'Silver', 'Black Smoke', 'Red Tick',
'Lilac Point', 'Red Merle', 'Tortie Point', 'Silver Tabby',
'Blue Cream', 'Yellow Brindle', 'Apricot', 'Green', 'Blue Point',
'Chocolate Point', 'Liver', 'Calico Point', 'Pink', 'Blue Tiger',
'Brown Tiger', 'Agouti', 'Silver Lynx Point', 'Blue Smoke',
'Liver Tick', 'Black Tiger', 'Orange Tiger', 'Cream Tiger',
'Gray Tiger', 'Ruddy'],
dtype='object'),
60)
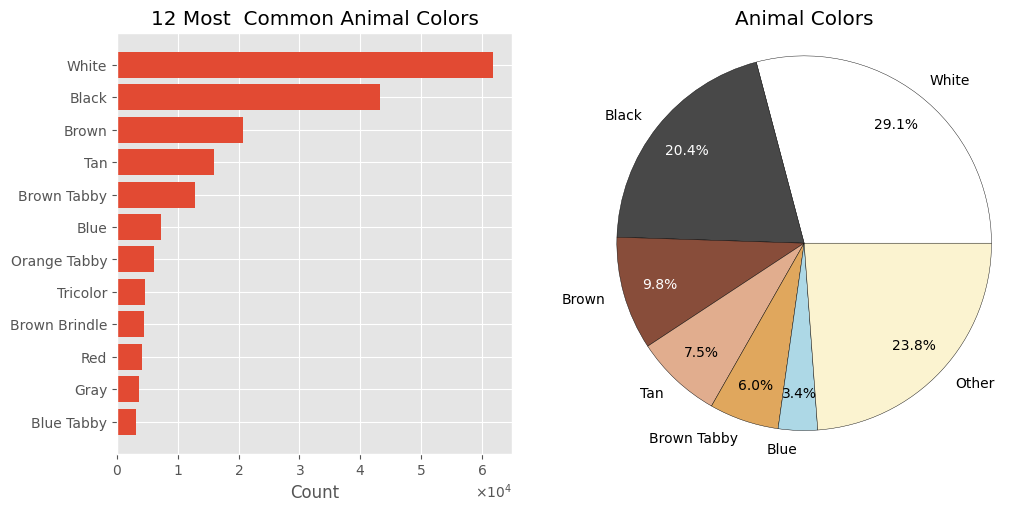
cols.head()
White 61893
Black 43306
Brown 20784
Tan 15939
Brown Tabby 12772
Name: count, dtype: int64
The Color feature is a categorical feature.
- The most common color combinations for animals at intake are black/white ⬛⬜, black ⬛, and brown 🟫.
- Considering all colors on an animal, a large portion are white ⬜, black ⬛, brown 🟫, or tan.
- The dataset contains 616 unique color combinations derived from 60 distinct colors.
fig, axes = plt.subplots(1, 2, figsize=(10, 5), layout='constrained', gridspec_kw={'wspace': 0.1})
ax = axes[0]
toshow = cols.iloc[:12]
ax.set_title('12 Most Common Animal Colors')
ax.set_xlabel('Count')
ax.ticklabel_format(axis='x', style='sci', scilimits=(0, 0), useMathText=True)
ax.barh(toshow.index, toshow)
ax.invert_yaxis()
ax = axes[1]
ax.set_title('Animal Colors')
t = 6
sumcol = cols.iloc[:t]
sumcol['Other'] = cols.iloc[t:].sum()
patches, _, autotexts = ax.pie(sumcol, labels=sumcol.index, autopct='%1.1f%%', pctdistance=0.8, frame=False,
colors=['white', '#484848', '#884d3a', '#e1ad8e', '#e0a75d', '#add8e6', '#fbf3d0'],
wedgeprops={"edgecolor":'black','linewidth': 0.3, 'antialiased': True})
ax.axis('equal')
autotexts[1].set_color('white')
autotexts[2].set_color('white')
⚕️ Intake Condition Feature
3. Finally, we’ll examine the Intake Condition feature using appropriate univariate descriptive statistics.
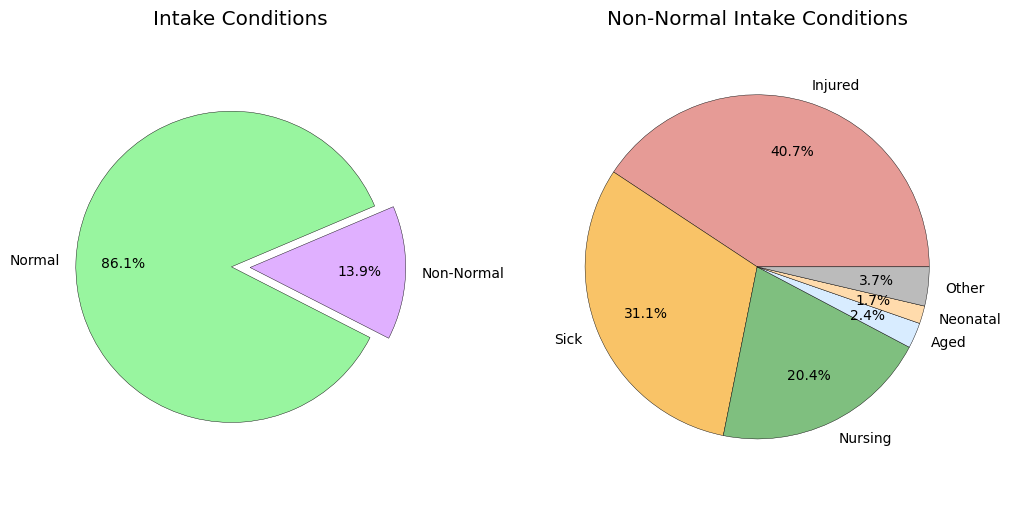
cond = intakes_df['Intake Condition'].value_counts()
cond
Intake Condition
Normal 119305
Injured 7841
Sick 5997
Nursing 3932
Aged 463
Neonatal 321
Other 245
Medical 174
Feral 125
Pregnant 103
Behavior 49
Space 4
Med Attn 3
Med Urgent 2
Panleuk 1
Name: count, dtype: int64
The Intake Condition feature is categorical.
- Statistics show that most animals are admitted in a Normal condition (86%).
- Of the remaining cases, animals are most frequently admitted as injured (40%), sick (31%), or nursing (20%).
normal = cond.loc['Normal']
nonnormal = cond.loc[cond.index != 'Normal']
fig, axes = plt.subplots(1, 2, figsize=(10, 5), layout='constrained', gridspec_kw={'wspace': 0.1})
# ax = axes[2]
# ax.set_title('Comparison of Non-Normal Intake Conditions')
# ax.set_xlabel('Count')
# ax.ticklabel_format(axis='x', style='sci', scilimits=(0, 0), useMathText=True)
# ax.barh(nonnormal.index, nonnormal)
# ax.invert_yaxis()
ax = axes[0]
ax.set_title('Intake Conditions')
patches, _, autotexts = ax.pie([normal, nonnormal.sum()], labels=['Normal', 'Non-Normal'], autopct='%1.1f%%', pctdistance=0.7, frame=False,
colors=['#98f59f', '#E0B0FF'], startangle=23, explode=(0, 0.12),
wedgeprops={"edgecolor":'black','linewidth': 0.3, 'antialiased': True})
ax.axis('equal')
ax = axes[1]
ax.set_title('Non-Normal Intake Conditions')
t = 5
sumcol = nonnormal.iloc[:t]
sumcol['Other'] = nonnormal.iloc[t:].sum()
patches, _, autotexts = ax.pie(sumcol, labels=sumcol.index, autopct='%1.1f%%', pctdistance=0.7, frame=False,
colors=['#e69b96', '#f9c367', '#7fbf7f', '#d8ecff', '#ffdbac', '#bbb'],
wedgeprops={"edgecolor":'black','linewidth': 0.3, 'antialiased': True})
ax.axis('equal');
🏠 Animal Origin
1. We’ll now select two features with a potential relationship (e.g., correlation) and describe this relationship using bivariate descriptive statistics.
sample = intakes_df[intakes_df['Animal Type'] == 'Other'].loc[:, 'Breed'].unique()
print(list(sample)[:20])
['Bat', 'Bat Mix', 'Hamster Mix', 'Raccoon', 'Raccoon Mix', 'Rabbit Sh Mix', 'Skunk Mix', 'Cinnamon', 'Rabbit Sh', 'Opossum', 'Skunk', 'Dutch/Angora-Satin', 'Fox', 'Rat', 'Guinea Pig Mix', 'Ferret', 'Cold Water', 'Rat Mix', 'Opossum Mix', 'Guinea Pig']
condtype = intakes_df.loc[:, ['Intake Type', 'Animal Type']].value_counts().unstack().fillna(0).astype(int).transpose()
cm = sns.light_palette(blue, as_cmap=True)
style = condtype.style.background_gradient(axis=1, cmap=cm).set_properties(**{'text-align':'center','font-size':'16px'})
style.map(lambda v: 'opacity: 30%;' if (v == 0) else None)
style.set_caption('Animal Types by Intake Type').set_table_styles([dict(
selector='caption', props=[('font-size', '150%'), ('text-align', 'left'), ('color', '#555')])])
| Intake Type | Abandoned | Euthanasia Request | Owner Surrender | Public Assist | Stray | Wildlife |
|---|---|---|---|---|---|---|
| Animal Type | ||||||
| Bird | 0 | 3 | 83 | 137 | 328 | 110 |
| Cat | 366 | 59 | 10586 | 1157 | 40205 | 0 |
| Dog | 347 | 183 | 17142 | 6735 | 53727 | 1 |
| Livestock | 0 | 0 | 1 | 1 | 22 | 0 |
| Other | 27 | 14 | 763 | 314 | 993 | 5261 |
Animals primarily arrive at the shelter through:
- For birds, cats, and dogs: being found as strays or being surrendered by their owners.
- For wild and exotic animals (categorized as “Other,” including bats, deer, snakes, frogs, etc.): being from the wild.
- A smaller number of animals are abandoned by their owners or are admitted with an euthanasia request.
The table visualization supports these observations. The values represent the frequency of each case, and the background color highlights the most frequent occurrences per row.
🚪 Animal Outcomes
2. Here, we will analyze the relationship between two features by employing bivariate descriptive statistics.
types = outcomes_df.loc[:, ['Outcome Subtype', 'Animal Type']].value_counts().unstack().fillna(0).astype(int).transpose().T
cm = sns.light_palette(blue, as_cmap=True)
style = types.style.background_gradient(axis=1, cmap=cm).set_properties(**{'text-align':'center','font-size':'16px'})
style.map(lambda v: 'opacity: 30%;' if (v == 0) else None)
style.set_caption('Animal Types by Outcome Type').set_table_styles([dict(
selector='caption', props=[('font-size', '150%'), ('text-align', 'left'), ('color', '#555')])])
| Animal Type | Bird | Cat | Dog | Livestock | Other |
|---|---|---|---|---|---|
| Outcome Subtype | |||||
| Aggressive | 0 | 4 | 565 | 0 | 1 |
| At Vet | 2 | 169 | 102 | 1 | 25 |
| Barn | 0 | 11 | 1 | 0 | 0 |
| Behavior | 0 | 0 | 159 | 0 | 0 |
| Court/Investigation | 0 | 0 | 38 | 0 | 0 |
| Customer S | 0 | 6 | 12 | 0 | 0 |
| Emer | 0 | 2 | 3 | 0 | 0 |
| Emergency | 0 | 3 | 4 | 0 | 0 |
| Enroute | 2 | 34 | 11 | 0 | 42 |
| Field | 0 | 6 | 91 | 0 | 0 |
| Foster | 38 | 7142 | 5249 | 10 | 101 |
| In Foster | 2 | 262 | 53 | 0 | 12 |
| In Kennel | 14 | 410 | 183 | 0 | 71 |
| In State | 0 | 0 | 11 | 0 | 0 |
| In Surgery | 0 | 14 | 12 | 0 | 1 |
| Medical | 11 | 87 | 78 | 0 | 151 |
| Offsite | 2 | 114 | 339 | 1 | 1 |
| Out State | 0 | 0 | 397 | 0 | 0 |
| Partner | 199 | 15723 | 16698 | 6 | 969 |
| Possible Theft | 0 | 2 | 14 | 0 | 0 |
| Prc | 0 | 4 | 8 | 0 | 0 |
| Rabies Risk | 0 | 100 | 95 | 0 | 3861 |
| SCRP | 0 | 3208 | 0 | 0 | 0 |
| Snr | 0 | 2935 | 0 | 0 | 0 |
| Suffering | 110 | 1821 | 891 | 1 | 686 |
| Underage | 1 | 1 | 0 | 0 | 34 |
Animals typically leave the shelter through:
- Most frequently, by adoption by new owners or by being placed in foster care.
- Some animals undergo euthanasia due to suffering.
- For wild and exotic animals (bats, deer, snakes, frogs, etc.), a common outcome is rabies risk.
- Stray cats are often neutralized through Shelter-Neuter-Return (SNR) programs or find homes via the Stray Cat Return Program (SCRP).
❓ Insights
💙 Intake and Outcome Type Dependency
Does an animal’s intake type influence its outcome type at the shelter? For simplicity, we’ll only consider animals that appear exactly once in each dataset (intake and outcome).
First, we create a merged dataset containing only animals with a single intake and a single outcome record.
sonce = intakes_df['Animal ID'].value_counts() == 1
in_wo_dupl = intakes_df.loc[intakes_df['Animal ID'].isin(sonce[sonce].index.values)]
sonce = outcomes_df['Animal ID'].value_counts() == 1
out_wo_dupl = outcomes_df.loc[outcomes_df['Animal ID'].isin(sonce[sonce].index.values)]
merged = intakes_df.merge(outcomes_df, on='Animal ID', suffixes=('_in', '_out'))
durs = merged['DateTime_out'] - merged['DateTime_in']
merged = merged[durs == abs(durs)]
counts = merged.loc[:, ['Intake Type', 'Outcome Type']].value_counts().unstack().fillna(0).astype(int)
cm = sns.light_palette(blue, as_cmap=True)
style = counts.style.background_gradient(axis=1, cmap=cm).set_properties(**{'text-align':'center','font-size':'16px'})
style.map(lambda v: 'opacity: 30%;' if (v == 0) else None)
style.set_caption('Outcome Types by Intake Type').set_table_styles([dict(
selector='caption', props=[('font-size', '150%'), ('text-align', 'left'), ('color', '#555')])])
| Outcome Type | Adoption | Died | Disposal | Euthanasia | Missing | Relocate | Return to Owner | Rto-Adopt | Transfer |
|---|---|---|---|---|---|---|---|---|---|
| Intake Type | |||||||||
| Abandoned | 438 | 4 | 4 | 5 | 0 | 0 | 57 | 9 | 224 |
| Euthanasia Request | 17 | 2 | 2 | 160 | 0 | 0 | 8 | 0 | 31 |
| Owner Surrender | 20666 | 184 | 13 | 802 | 11 | 0 | 1722 | 338 | 7987 |
| Public Assist | 1759 | 51 | 57 | 515 | 3 | 0 | 6712 | 188 | 1244 |
| Stray | 49253 | 910 | 133 | 2892 | 63 | 8 | 18973 | 771 | 30324 |
| Wildlife | 7 | 134 | 396 | 4406 | 2 | 14 | 3 | 0 | 51 |
It’s clear that the intake type significantly impacts the outcome type.
- Animals classified as Abandoned, Owner Surrender, or Stray at intake are frequently Adopted.
- Animals entering via Public Assist are often Returned to Owner, or adopted in about a fifth of cases.
- Wildlife and animals received through an Euthanasia Request are frequently euthanized.
🐥 Animal Age at Adoption
Does an animal’s age play a role in its adoption?
We’ll categorize outcomes into Adopted and Not Adopted.
adopted = outcomes_df.loc[outcomes_df['Outcome Type'] == 'Adoption', 'Age upon Outcome']/365
notadopted = outcomes_df.loc[outcomes_df['Outcome Type'] != 'Adoption', 'Age upon Outcome'].dropna()/365
fig, (ax1, ax2) = plt.subplots(nrows=1, ncols=2, figsize=(9, 4), sharey=True, layout='constrained')
fig.suptitle('Adoption Rate by Age', fontsize=16)
ax1.xaxis.grid(True)
ax1.set_title('Full range', fontsize=12)
ax1.set_xlabel('Age [years]')
ax1.set_yticks([1, 2], labels=['Adopted', 'Not Adopted'])
parts = ax1.violinplot([adopted.astype(float), notadopted.astype(float)], [1, 2],
widths=0.6, points=100, vert=False, showmeans=True)
parts['bodies'][0].set_facecolor(red)
parts['bodies'][1].set_facecolor(blue)
for part in ['cbars', 'cmaxes', 'cmins', 'cmeans']:
parts[part].set_color([red, blue])
ax1.invert_yaxis()
ax2.xaxis.grid(True)
ax2.set_title('Age < 6 (focused)', fontsize=12)
ax2.set_xlabel('Age [years]')
parts = ax2.violinplot([adopted.astype(float), notadopted.astype(float)], [1, 2],
widths=0.8, points=150, vert=False, showmeans=True)
parts['bodies'][0].set_facecolor(red)
parts['bodies'][1].set_facecolor(blue)
for part in ['cbars', 'cmaxes', 'cmins', 'cmins']:
parts[part].set_color([red, blue])
ax2.set_xlim(left=-.4, right=6);
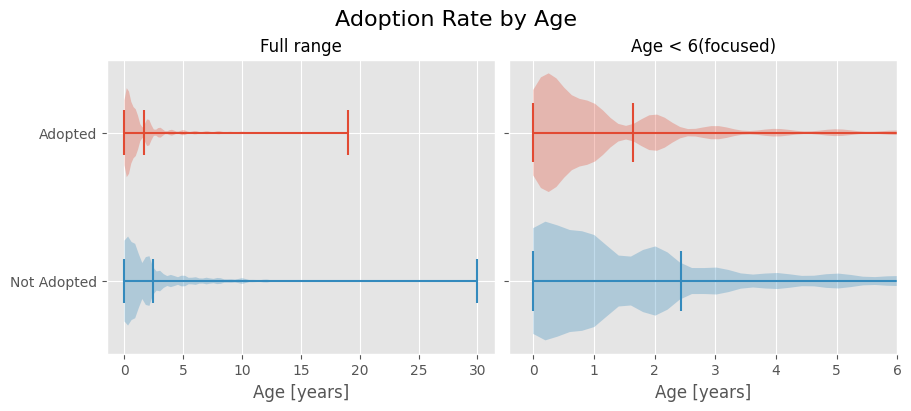
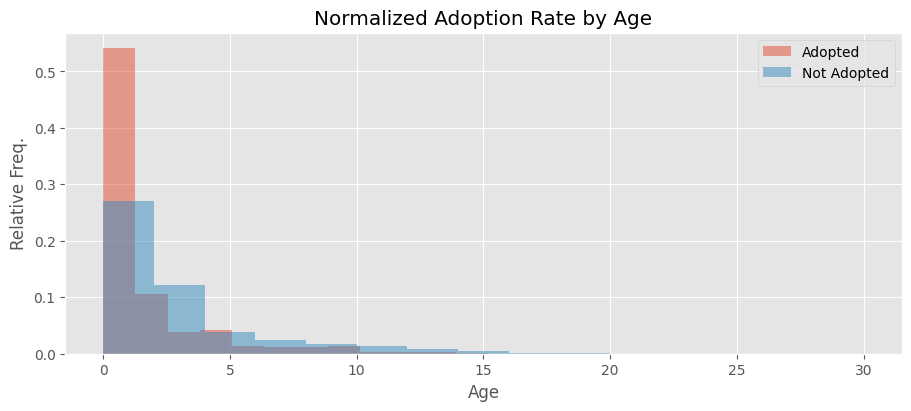
This visualization reveals that non-adopted animals are relatively well-represented across higher age categories, while adopted animals tend to be younger.
The difference becomes more apparent after normalization, as there are more non-adopted animals in the dataset. This suggests that adoption is more common for younger animals.
fig, ax = plt.subplots(nrows=1, ncols=1, figsize=(9, 4), layout='constrained')
ax.set_title('Normalized Adoption Rate by Age')
ax.hist(adopted, 15, alpha=0.5, color=(226/255, 74/255, 51/255), label='Adopted', density=True)
ax.hist(notadopted, 15, alpha=0.5, label='Not Adopted', color=(52/255, 138/255, 189/255), density=True)
ax.set_xlabel('Age')
ax.set_ylabel('Relative Freq.')
ax.legend();
📈 Animal Intake Trends
Is the animal intake consistent throughout the year, or are there periods of higher or lower load?
time_ser = intakes_df['DateTime'].dt.strftime('%Y-%m').value_counts().sort_index()
fig, (ax1, ax2) = plt.subplots(2, 1, figsize=(9, 8), layout='constrained', gridspec_kw={'hspace': 0.1})
ax1.plot(time_ser)
ticks = list(time_ser.index)[3::12]
ticklabels = list(time_ser.index)[3+5::12]
ax1.set_xticks(ticks)
ax1.set_xticklabels(f"{tick[:-3]}" for tick in ticks)
ax1.set_title('Intakes over Time')
ax1.set_ylim(ymin=0)
ax1.set_xlabel('Year')
ax1.set_ylabel('Intake Count')
ax1.grid(which='major', axis='x', color='grey', linestyle='dotted', linewidth=0.6)
dt = intakes_df[['DateTime']].copy()
dt['Year'] = dt['DateTime'].dt.year
dt['Month'] = dt['DateTime'].dt.month
dt = dt.drop('DateTime', axis=1)
dt = dt.value_counts().groupby(['Year', 'Month']).sum()
means = dt.groupby('Month').mean()
bar_labels=['Jan', 'Feb', 'Mar', 'Apr', 'May', 'Jun', 'Jul', 'Aug', 'Sep', 'Oct', 'Nov', 'Dec']
(main, second, rest) = (red, '#ea8070', '#f3b6ad')
bar_colors = [rest, rest, rest, rest, main, main, second, second, second, second, rest, rest]
p = ax2.bar(means.index, means, width=0.6, color=bar_colors)
ax2.set_title('Average Monthly Intakes')
ax2.set_xticks(range(1, 13), labels=bar_labels)
ax2.set_xlabel('Month')
ax2.bar_label(p, fmt='%i', padding=-20, color='w')
ax2.set_yticks([])
ax2.set_ylim(ymin=0)
ax2.set_ylabel('Average Intake Count')
ax2.grid(visible=False, which='major', axis='both')
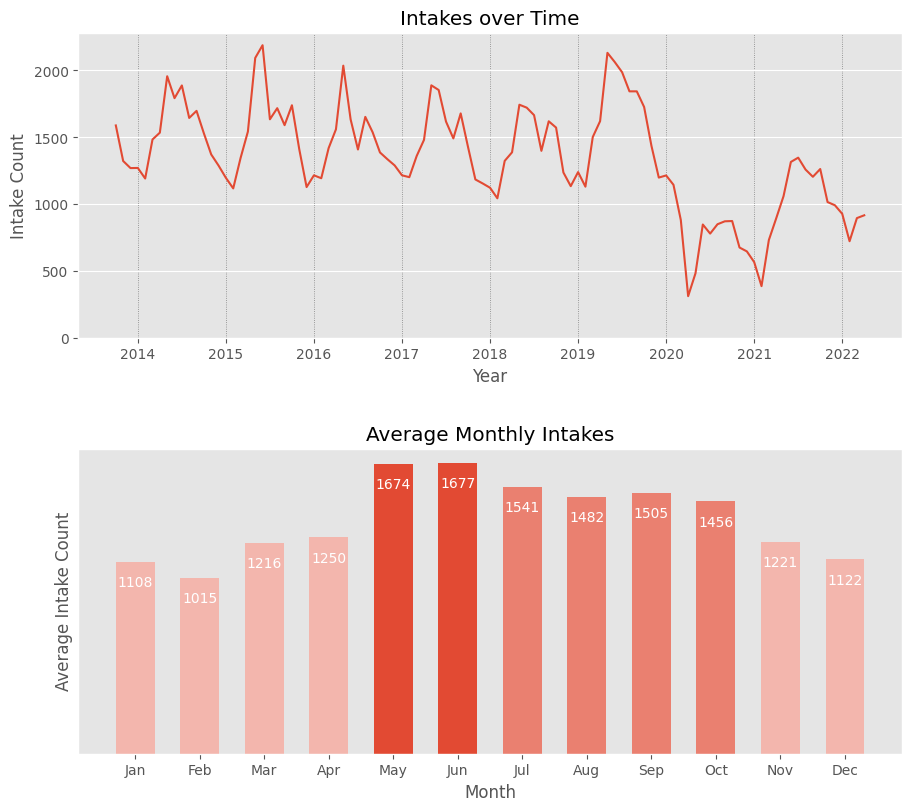
The first graph shows that there are specific periods of the year when animal intake is significantly higher. The second graph reveals that this period typically lasts from May to October, with an average intake of about 500 more animals per month compared to other months. Intake is lowest during winter and spring.
Note: The first graph also shows a sharp drop in intakes at the end of 2019, likely due to the COVID-19 pandemic. Since then, intake frequency has been slowly rising.
💔 Animal Sterilization Status
How does the sterilization status of animals change from intake to outcome?
counts = merged.loc[:, ['Sterile_in', 'Sterile_out']].value_counts()
scounts = counts.reset_index().reindex([2, 0, 1]).reset_index(drop=True)
cnt_nst_in = scounts.loc[scounts['Sterile_in'] == False, 'count'].sum()
cnt_st_in = scounts.loc[scounts['Sterile_in'] == True, 'count'].sum()
scounts.loc[:2, 'frac'] = scounts.loc[:2, 'count']/cnt_nst_in
scounts.loc[2:, 'frac'] = scounts.loc[2:, 'count']/cnt_st_in
scounts['frac'] = scounts['frac']
cm = sns.light_palette(blue, as_cmap=True)
style = scounts.style.set_properties(**{'text-align':'center','font-size':'16px'})
style.set_caption('Animal Types by Origin').set_table_styles([dict(
selector='caption', props=[('font-size', '150%'), ('text-align', 'right'), ('color', '#555')])])
style.format(precision=2)
| Sterile_in | Sterile_out | count | frac | |
|---|---|---|---|---|
| 0 | False | False | 32705 | 0.34 |
| 1 | False | True | 63245 | 0.66 |
| 2 | True | True | 45695 | 1.00 |
counts = merged.loc[:, ['Sterile_in', 'Sterile_out']].value_counts()
st_in, nst_in, st_out, nst_out = ('Sterile (in)', 'Non Sterile (in)', 'Sterile (out)', 'Non Sterile (out)')
cdict = {st_in: {st_out: counts.loc[True, True]},
nst_in: {st_out: counts.loc[False, True], nst_out: counts.loc[False, False]}}
ax = alluvial.plot(cdict, figsize=(6, 4), fontname='Monospace', color_side=0,
alpha=0.9, colors=[red, blue], src_label_override=[st_in, nst_in], dst_label_override=[st_out, nst_out],
disp_width=True, wdisp_sep=3*' ', width_in=False, v_gap_frac=0.1)
ax.set_title('Animal Sex Transitions')
left, width = .25, .5
bottom, height = .25, .5
right = left + width
top = bottom + height
ax.text(0.5, 0.55, 'neutering', fontstyle='oblique',
horizontalalignment='center', verticalalignment='center',
transform=ax.transAxes, color='w', fontsize=14);
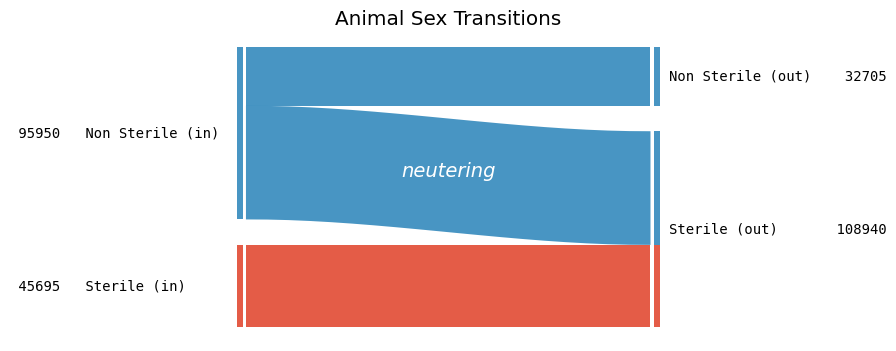
Again, for simplicity, we only consider animals that entered and left the shelter exactly once.
The visualization clearly shows that a large proportion (66%) of incoming non-sterile animals undergo sterilization at the shelter. Notably, no animal managed to reverse the procedure!
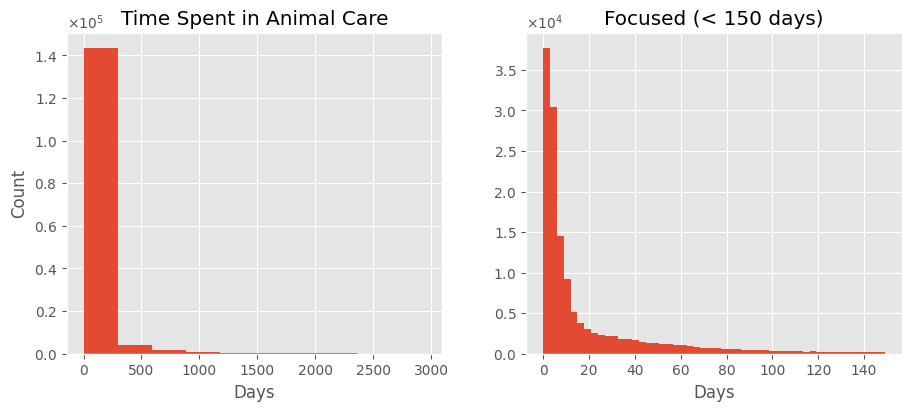
⏳ Time Spent in Shelter
How long do animals typically stay at the shelter?
durs = (merged['DateTime_out'] - merged['DateTime_in']).dt.days
print_univar_num_stats(durs)
None statistics
min: 0
max: 2948
mean: 63.78191485852635
median: 7.0
range: 2948
lower quartile: 3.0
upper quartile: 34.0
IQR: 31.0
variance: 40540.078983925494
std. variation: 201.345670387832
skewness: 6.139468243278622
kurtosis: 46.946690264259104
fig, (ax1, ax2) = plt.subplots(nrows=1, ncols=2, figsize=(9, 4), sharey=False, layout='constrained', gridspec_kw={'wspace': 0.1})
ax1.hist(durs)
ax1.set_title('Time Spent in Animal Care')
ax1.set_xlabel('Days')
ax1.set_ylabel('Count')
ax1.ticklabel_format(axis='y', style='sci', scilimits=(0,0), useMathText=True)
ax2.hist(durs[durs < 150], bins=50)
ax2.set_title('Focused (< 150 days)')
ax2.set_xlabel('Days')
ax2.ticklabel_format(axis='y', style='sci', scilimits=(0,0), useMathText=True)
durs.sort_values(ascending=False).head(5) / 360
176625 8.188889
90634 8.163889
31455 8.077778
171654 8.072222
177941 8.052778
dtype: float64
Considering only animals that have already left the shelter, most stays are under 3 months, often even less than a month. While some animals stay for up to 8 years, the average duration is 63 days, with a median of just 7 days.
Note: The y-axis scale in the right histogram is different.
💉 Euthanasia Frequency
Is the frequency of euthanasias constant, or are there periods when they are more common?
counts = merged.loc[merged['Outcome Type'] == 'Euthanasia', 'DateTime_out']
fig, (ax1, ax2) = plt.subplots(nrows=2, ncols=1, figsize=(9, 8), sharey=False, layout='constrained', gridspec_kw={'hspace': 0.1})
ax1.set_title('Frequency of Euthanasia over Time')
time_ser = counts.dt.strftime('%Y-%m').value_counts().sort_index()
ax1.plot(time_ser, label='Real data', alpha=0.4)
ax1.plot(time_ser.rolling(window=12).mean(), label='Rolling average [annual]', linewidth=2.2)
ticks = list(time_ser.index)[3::12]
ticklabels = list(time_ser.index)[3+5::12]
ax1.set_xticks(ticks)
ax1.set_xticklabels(f"{tick[:-3]}" for tick in ticks)
ax1.set_ylim(ymin=0)
ax1.set_xlabel('Year')
ax1.set_ylabel('Euthanasia Procedure Count')
ax1.legend()
dt = merged.loc[merged['Outcome Type'] == 'Euthanasia', ['DateTime_out']].copy()
dt['Year'] = dt['DateTime_out'].dt.year
dt['Month'] = dt['DateTime_out'].dt.month
dt = dt.drop('DateTime_out', axis=1)
dt = dt.value_counts().groupby(['Year', 'Month']).sum()
means = dt.groupby('Month').mean()
bar_labels=['Jan', 'Feb', 'Mar', 'Apr', 'May', 'Jun', 'Jul', 'Aug', 'Sep', 'Oct', 'Nov', 'Dec']
(main, second, rest) = (red, '#ea8070', '#f3b6ad')
bar_colors = [rest, rest, main, second, second, second, rest, rest, rest, rest, rest, rest]
p = ax2.bar(means.index, means, width=0.6, color=bar_colors)
ax2.set_title('Frequency of Euthanasia each Month')
ax2.set_xticks(range(1, 13), labels=bar_labels)
ax2.set_xlabel('Month')
ax2.bar_label(p, fmt='%i', padding=-20, color='w')
ax2.set_yticks([])
ax2.set_ylim(ymin=0)
ax2.set_ylabel('Average Intake Count')
ax2.grid(visible=False, which='major', axis='both')
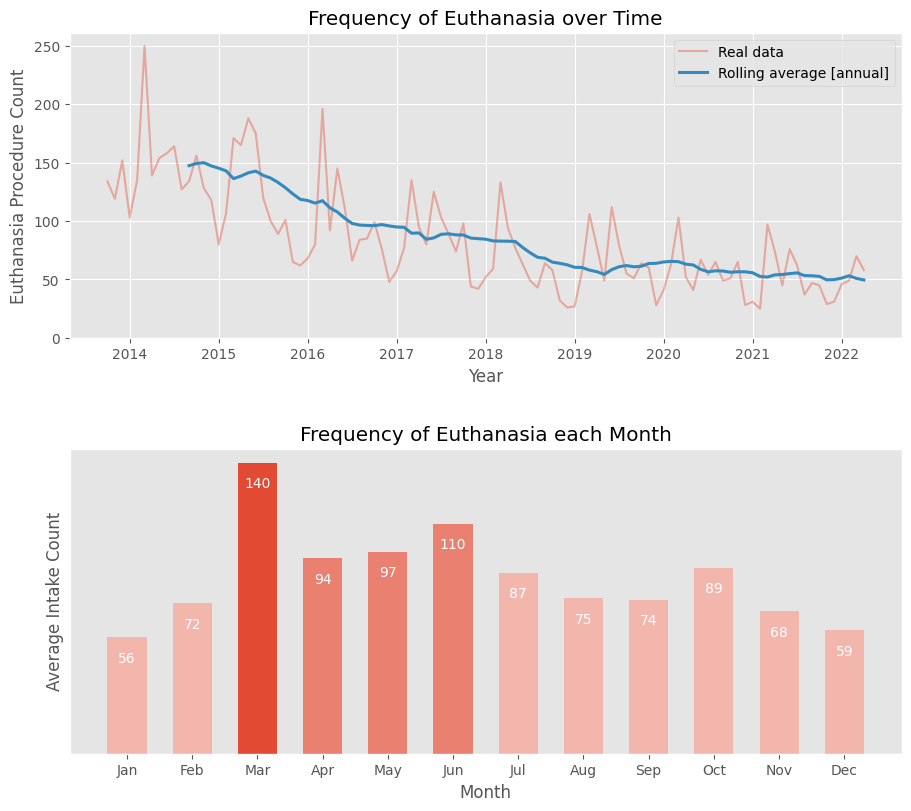
The first graph, showing the moving average, indicates a decline in euthanasia procedures since 2013, dropping from an average of 150 per month to around 50.
Despite this decline, the overall monthly averages still appear high due to the influence of older data. The highest frequency of procedures occurs in March, with an average of 140 animals euthanized. The average remains around 100 for the next three months. For the rest of the year, the number of procedures is generally below 90 per month.