Here’s a breakdown of the features used in this analysis:
- Year: The year of observation.
- Status: Indicates whether the country is Developed or Developing.
- Life expectancy: Life expectancy in years – this is our target variable to predict.
- Adult Mortality: Adult mortality rate (probability of dying between 15 and 60 years per 1,000 population).
- infant deaths: Number of infant deaths per 1,000 population.
- Alcohol: Recorded per capita consumption (15+) of pure alcohol (in liters).
- percentage expenditure: Percentage of gross domestic product (GDP) spent on health per capita (%).
- Hepatitis B: Hepatitis B (HepB) immunization coverage among 1-year-olds (%).
- Measles: Number of reported measles cases per 1,000 population.
- BMI: Average Body Mass Index of the entire population.
- under-five deaths: Number of under-five deaths per 1,000 population.
- Polio: Polio (Pol3) immunization coverage among 1-year-olds (%).
- Total expenditure: Government health expenditure as a percentage of total government expenditure (%).
- Diphtheria: Diphtheria, Tetanus, and Pertussis (DTP3) immunization coverage among 1-year-olds (%).
- HIV/AIDS: Deaths per 1,000 live births due to HIV/AIDS (0-4 years).
- GDP: Gross Domestic Product per capita (in USD).
- Population: Country’s population.
- thinness 1-19 years: Prevalence of thinness among children aged 10-19 (BMI less than 2 standard deviations below the median) (%).
- thinness 5-9 years: Prevalence of thinness among children aged 5-9 (BMI less than 2 standard deviations below the median) (%).
- Income composition of resources: Human Development Index (HDI) based on income composition of resources (index ranging from 0 to 1).
- Schooling: Number of years of schooling (years).
📦 Library Imports
This section handles the necessary imports, sets a random seed for reproducibility, and configures plotting styles.
from itertools import chain
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import matplotlib.colors as mcolors
import missingno as msno
import pycountry_convert as pcc
from matplotlib.ticker import MaxNLocator
from matplotlib.ticker import FormatStrFormatter
from sklearn.model_selection import train_test_split
from sklearn.model_selection import ParameterGrid
from sklearn.metrics import mean_squared_error as mse
from sklearn.metrics import mean_absolute_error as mae
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import OneHotEncoder
from sklearn.preprocessing import PolynomialFeatures
from sklearn.preprocessing import MinMaxScaler
from sklearn.preprocessing import MaxAbsScaler
from sklearn.preprocessing import StandardScaler
from sklearn.preprocessing import StandardScaler
from sklearn.preprocessing import FunctionTransformer
from sklearn.tree import DecisionTreeRegressor
from sklearn.base import RegressorMixin
from sklearn.base import BaseEstimator
from sklearn.base import TransformerMixin
from sklearn.linear_model import Ridge
from sklearn.neighbors import KNeighborsRegressor
from sklearn.utils import resample
from sklearn.impute import SimpleImputer
from sklearn.impute import KNNImputer
from sklearn.compose import ColumnTransformer
np.random.seed(42)
plt.style.use('ggplot')
red = np.array((226/255, 74/255, 51/255))
blue = np.array((52/255, 138/255, 189/255))
grey = np.array((100/255, 100/255, 100/255))
cobalt = np.array((0/255, 71/255, 171/255))
main_color = cobalt
⚙️ Data Preprocessing
This section covers loading the dataset, performing initial exploratory data analysis, visualizing features, and splitting the data into training, validation, and test sets.
📚 Dataset Loading
raw_data = pd.read_csv('data.csv')
raw_data.head().T
| 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| Country | Afghanistan | Afghanistan | Afghanistan | Afghanistan | Afghanistan |
| Year | 2015 | 2014 | 2013 | 2012 | 2011 |
| Status | Developing | Developing | Developing | Developing | Developing |
| Life expectancy | 65.0 | 59.9 | 59.9 | 59.5 | 59.2 |
| Adult Mortality | 263.0 | 271.0 | 268.0 | 272.0 | 275.0 |
| infant deaths | 62 | 64 | 66 | 69 | 71 |
| Alcohol | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 |
| percentage expenditure | 71.279624 | 73.523582 | 73.219243 | 78.184215 | 7.097109 |
| Hepatitis B | 65.0 | 62.0 | 64.0 | 67.0 | 68.0 |
| Measles | 1154 | 492 | 430 | 2787 | 3013 |
| BMI | 19.1 | 18.6 | 18.1 | 17.6 | 17.2 |
| under-five deaths | 83 | 86 | 89 | 93 | 97 |
| Polio | 6.0 | 58.0 | 62.0 | 67.0 | 68.0 |
| Total expenditure | 8.16 | 8.18 | 8.13 | 8.52 | 7.87 |
| Diphtheria | 65.0 | 62.0 | 64.0 | 67.0 | 68.0 |
| HIV/AIDS | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| GDP | 584.25921 | 612.696514 | 631.744976 | 669.959 | 63.537231 |
| Population | 33736494.0 | 327582.0 | 31731688.0 | 3696958.0 | 2978599.0 |
| thinness 1-19 years | 17.2 | 17.5 | 17.7 | 17.9 | 18.2 |
| thinness 5-9 years | 17.3 | 17.5 | 17.7 | 18.0 | 18.2 |
| Income composition of resources | 0.479 | 0.476 | 0.47 | 0.463 | 0.454 |
| Schooling | 10.1 | 10.0 | 9.9 | 9.8 | 9.5 |
💡 Dataset Overview
pd.DataFrame([raw_data.count(), raw_data.nunique(), raw_data.dtypes], index=['Non-Null Count', 'Unique Values', 'Dtype']).T
| Non-Null Count | Unique Values | Dtype | |
|---|---|---|---|
| Country | 2718 | 183 | object |
| Year | 2718 | 16 | int64 |
| Status | 2718 | 2 | object |
| Life expectancy | 2718 | 359 | float64 |
| Adult Mortality | 2718 | 423 | float64 |
| infant deaths | 2718 | 195 | int64 |
| Alcohol | 2564 | 1055 | float64 |
| percentage expenditure | 2718 | 2185 | float64 |
| Hepatitis B | 2188 | 87 | float64 |
| Measles | 2718 | 909 | int64 |
| BMI | 2692 | 600 | float64 |
| under-five deaths | 2718 | 239 | int64 |
| Polio | 2700 | 73 | float64 |
| Total expenditure | 2529 | 792 | float64 |
| Diphtheria | 2700 | 81 | float64 |
| HIV/AIDS | 2718 | 197 | float64 |
| GDP | 2317 | 2317 | float64 |
| Population | 2116 | 2110 | float64 |
| thinness 1-19 years | 2692 | 194 | float64 |
| thinness 5-9 years | 2692 | 200 | float64 |
| Income composition of resources | 2576 | 613 | float64 |
| Schooling | 2576 | 173 | float64 |
raw_data.describe().T
| count | mean | std | min | 25% | 50% | 75% | max | |
|---|---|---|---|---|---|---|---|---|
| Year | 2718.0 | 2.007114e+03 | 4.537979e+00 | 2000.00000 | 2003.000000 | 2.007000e+03 | 2.011000e+03 | 2.015000e+03 |
| Life expectancy | 2718.0 | 6.920453e+01 | 9.612530e+00 | 36.30000 | 63.100000 | 7.220000e+01 | 7.580000e+01 | 8.900000e+01 |
| Adult Mortality | 2718.0 | 1.644323e+02 | 1.255128e+02 | 1.00000 | 73.250000 | 1.420000e+02 | 2.270000e+02 | 7.230000e+02 |
| infant deaths | 2718.0 | 3.082524e+01 | 1.217866e+02 | 0.00000 | 0.000000 | 3.000000e+00 | 2.200000e+01 | 1.800000e+03 |
| Alcohol | 2564.0 | 4.672512e+00 | 4.051664e+00 | 0.01000 | 0.990000 | 3.820000e+00 | 7.832500e+00 | 1.787000e+01 |
| percentage expenditure | 2718.0 | 7.570717e+02 | 2.007472e+03 | 0.00000 | 5.832385 | 6.768701e+01 | 4.468877e+02 | 1.947991e+04 |
| Hepatitis B | 2188.0 | 8.088483e+01 | 2.501008e+01 | 1.00000 | 77.000000 | 9.200000e+01 | 9.700000e+01 | 9.900000e+01 |
| Measles | 2718.0 | 2.371000e+03 | 1.117424e+04 | 0.00000 | 0.000000 | 1.800000e+01 | 3.720000e+02 | 2.121830e+05 |
| BMI | 2692.0 | 3.831434e+01 | 1.995480e+01 | 1.00000 | 19.200000 | 4.345000e+01 | 5.610000e+01 | 7.760000e+01 |
| under-five deaths | 2718.0 | 4.276748e+01 | 1.657044e+02 | 0.00000 | 0.000000 | 4.000000e+00 | 2.800000e+01 | 2.500000e+03 |
| Polio | 2700.0 | 8.252815e+01 | 2.329438e+01 | 3.00000 | 77.000000 | 9.300000e+01 | 9.700000e+01 | 9.900000e+01 |
| Total expenditure | 2529.0 | 5.943606e+00 | 2.488801e+00 | 0.37000 | 4.260000 | 5.730000e+00 | 7.530000e+00 | 1.760000e+01 |
| Diphtheria | 2700.0 | 8.213593e+01 | 2.384957e+01 | 2.00000 | 78.000000 | 9.300000e+01 | 9.700000e+01 | 9.900000e+01 |
| HIV/AIDS | 2718.0 | 1.788263e+00 | 5.221587e+00 | 0.10000 | 0.100000 | 1.000000e-01 | 8.000000e-01 | 5.060000e+01 |
| GDP | 2317.0 | 7.646460e+03 | 1.445559e+04 | 1.68135 | 459.291200 | 1.741143e+03 | 6.337883e+03 | 1.191727e+05 |
| Population | 2116.0 | 1.261063e+07 | 6.238395e+07 | 34.00000 | 182922.000000 | 1.365022e+06 | 7.383590e+06 | 1.293859e+09 |
| thinness 1-19 years | 2692.0 | 4.892236e+00 | 4.434584e+00 | 0.10000 | 1.600000 | 3.400000e+00 | 7.200000e+00 | 2.770000e+01 |
| thinness 5-9 years | 2692.0 | 4.925149e+00 | 4.522269e+00 | 0.10000 | 1.600000 | 3.400000e+00 | 7.300000e+00 | 2.860000e+01 |
| Income composition of resources | 2576.0 | 6.266968e-01 | 2.133229e-01 | 0.00000 | 0.492000 | 6.790000e-01 | 7.810000e-01 | 9.380000e-01 |
| Schooling | 2576.0 | 1.199608e+01 | 3.364109e+00 | 0.00000 | 10.100000 | 1.230000e+01 | 1.430000e+01 | 2.070000e+01 |
✂️ Data Splitting
y_data = raw_data.loc[:, 'Life expectancy']
X_data = raw_data.drop('Life expectancy', axis=1)
test_ratio = 0.3
val_ratio = 0.2
X_train_val, X_test, y_train_val, y_test = train_test_split(X_data, y_data, test_size=test_ratio)
X_train, X_val, y_train, y_val = train_test_split(X_train_val, y_train_val, test_size=val_ratio/(1-test_ratio))
split_df = pd.DataFrame([X_train.shape[0], X_val.shape[0], X_test.shape[0]], index=['Train', 'Val', 'Test'], columns=['Size'])
split_df['Relative size'] = split_df['Size'] / split_df['Size'].sum()
split_df['Relative size'] = split_df['Relative size'].round(3)
split_df
| Size | Relative size | |
|---|---|---|
| Train | 1358 | 0.5 |
| Val | 544 | 0.2 |
| Test | 816 | 0.3 |
🕵️ Exploratory Data Analysis
This exploratory analysis focuses on:
- Features with missing values.
- The scale of individual features.
- The correlation of individual features with the target variable.
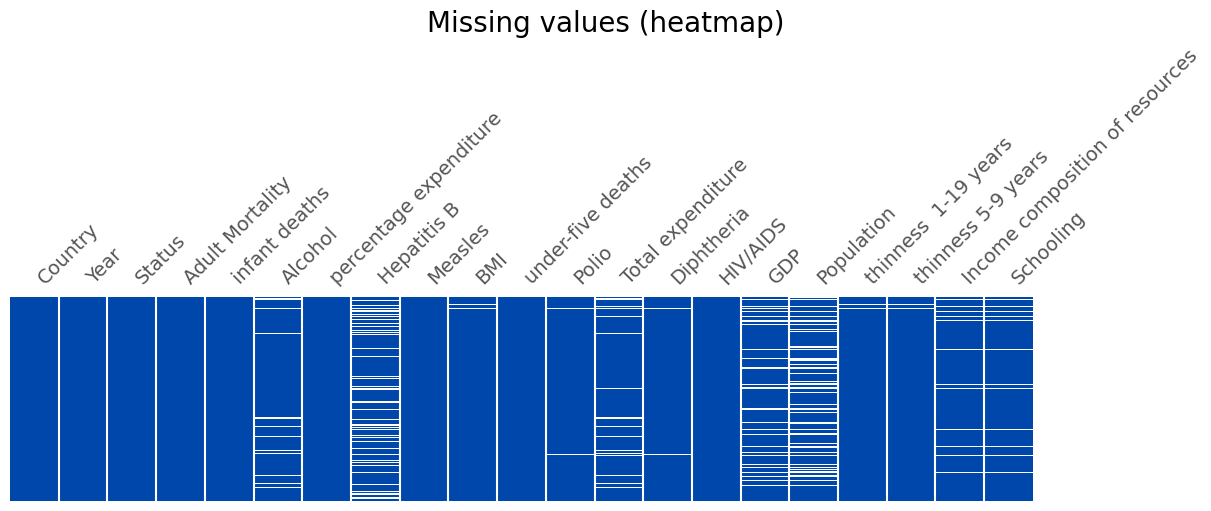
❓ Missing Values
fig, ax = plt.subplots(1, 1, figsize=(12, 5), layout='constrained')
fig.suptitle('Missing values (heatmap)', fontsize=20)
msno.matrix(X_train, fontsize=14, sparkline=False, ax=ax, color=main_color)
ax.get_yaxis().set_visible(False)
fig, ax = plt.subplots(1, 1, figsize=(12, 8), layout='constrained')
fig.suptitle('Missing values (counts)', fontsize=20)
missings = X_train.isna().sum().sort_values()
bars = ax.barh(missings.index, missings, color=main_color)
ax.set_xlabel('Count', fontsize=14)
ax.tick_params(axis='both', which='major', labelsize=12)
for bar, count in zip(bars, missings):
ax.text(bar.get_width()+2, bar.get_y() + bar.get_height() / 2, f'{count}', va='center', fontsize=10)
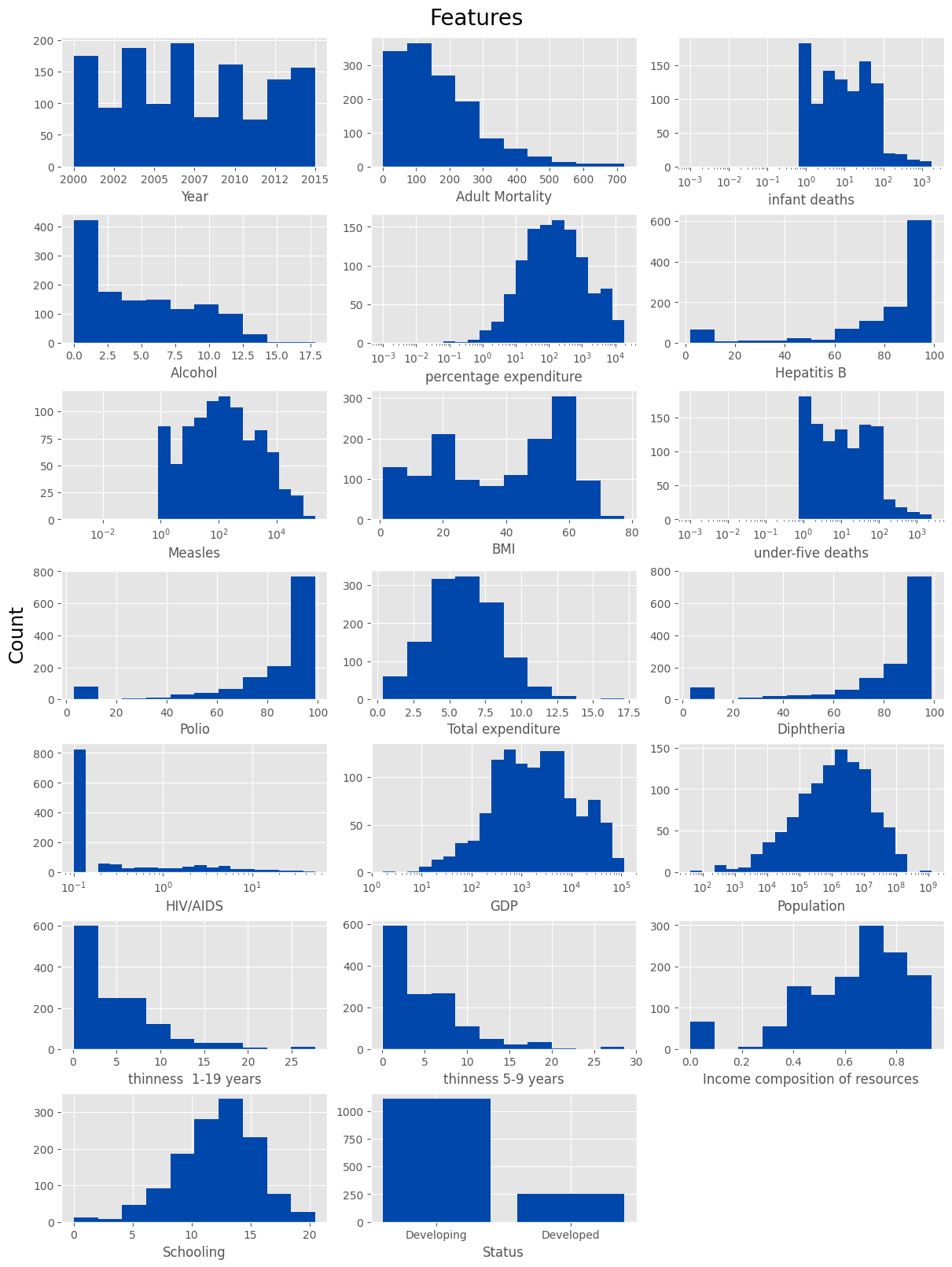
✨ Features
Here we examine the distribution of individual features. Features with a logarithmic scale are processed separately.
cat_columns = ['Status', 'Country']
num_columns = ['Year', 'Adult Mortality', 'infant deaths', 'Alcohol', 'percentage expenditure', 'Hepatitis B',
'Measles', 'BMI', 'under-five deaths', 'Polio', 'Total expenditure', 'Diphtheria', 'HIV/AIDS', 'GDP', 'Population',
'thinness 1-19 years', 'thinness 5-9 years', 'Income composition of resources', 'Schooling']
logscaled_columns = ['infant deaths', 'percentage expenditure', 'Measles', 'under-five deaths', 'HIV/AIDS', 'GDP', 'Population']
fig, axes = plt.subplots(7, 3, figsize=(12, 16), layout='constrained')
fig.suptitle('Features', size=20)
fig.supylabel('Count', size=18)
for col, ax in zip(num_columns, fig.axes):
data = X_train[col].dropna()
if col in logscaled_columns:
hist, bins = np.histogram(data, bins=20)
logbins = np.logspace(np.log10(max(bins[0], 0.001)), np.log10(bins[-1]), len(bins))
ax.set_xscale('log')
ax.hist(data, bins=logbins, color=main_color)
else:
ax.hist(data, color=main_color)
if col == 'Year':
ax.xaxis.set_major_formatter(FormatStrFormatter('%d'))
ax.set_xlabel(col)
ax = axes[6][1]
states = X_train['Status'].value_counts()
ax.bar(states.index, states, color=main_color)
ax.set_xlabel('Status')
axes[6][2].axis('off')
plt.show()
print('List of countries:')
print(', '.join(X_train['Country'].unique()))
List of countries:
Rwanda, Ukraine, Micronesia (Federated States of), Grenada, Montenegro, Sierra Leone, Mali, Lebanon, Cabo Verde, Tajikistan, Georgia, Spain, Algeria, Thailand, Fiji, Azerbaijan, United States of America, Cambodia, Togo, Russian Federation, France, Kyrgyzstan, Seychelles, Estonia, Gabon, Switzerland, Barbados, Nigeria, Turkmenistan, Comoros, Uruguay, Côte d'Ivoire, Samoa, Kazakhstan, Yemen, Belize, Iran (Islamic Republic of), Benin, Sri Lanka, Belgium, Italy, Zambia, Lithuania, Sudan, Burundi, Republic of Moldova, Papua New Guinea, Ethiopia, Sweden, Serbia, Jamaica, Denmark, Venezuela (Bolivarian Republic of), Afghanistan, South Africa, Democratic People's Republic of Korea, Maldives, Albania, Cyprus, India, Philippines, Tunisia, Saint Lucia, South Sudan, Zimbabwe, Angola, Nepal, Viet Nam, Croatia, Somalia, Brunei Darussalam, Antigua and Barbuda, Mauritius, Niger, Syrian Arab Republic, Morocco, Burkina Faso, New Zealand, United Kingdom of Great Britain and Northern Ireland, Guatemala, Malta, Republic of Korea, Honduras, Sao Tome and Principe, Trinidad and Tobago, Chile, Ghana, Cameroon, Oman, Ireland, Indonesia, Tonga, Hungary, Kenya, Malawi, Luxembourg, El Salvador, Slovenia, Jordan, Haiti, Namibia, Netherlands, Eritrea, Guyana, Argentina, Timor-Leste, Costa Rica, Guinea, Senegal, Germany, China, Uzbekistan, Ecuador, Suriname, Czechia, Equatorial Guinea, Greece, Belarus, Malaysia, Israel, Bosnia and Herzegovina, Bangladesh, Latvia, Pakistan, Myanmar, Iceland, Mauritania, Turkey, Mexico, Congo, Lao People's Democratic Republic, Austria, Mozambique, United Republic of Tanzania, Saudi Arabia, Singapore, Swaziland, United Arab Emirates, Qatar, The former Yugoslav republic of Macedonia, Bahrain, Nicaragua, Panama, Bhutan, Poland, Bolivia (Plurinational State of), Australia, Norway, Peru, Djibouti, Dominican Republic, Portugal, Armenia, Madagascar, Bahamas, Chad, Saint Vincent and the Grenadines, Mongolia, Botswana, Libya, Vanuatu, Bulgaria, Egypt, Canada, Guinea-Bissau, Lesotho, Democratic Republic of the Congo, Brazil, Kiribati, Iraq, Gambia, Slovakia, Kuwait, Japan, Paraguay, Uganda, Colombia, Finland, Liberia, Romania, Cuba, Solomon Islands, Central African Republic
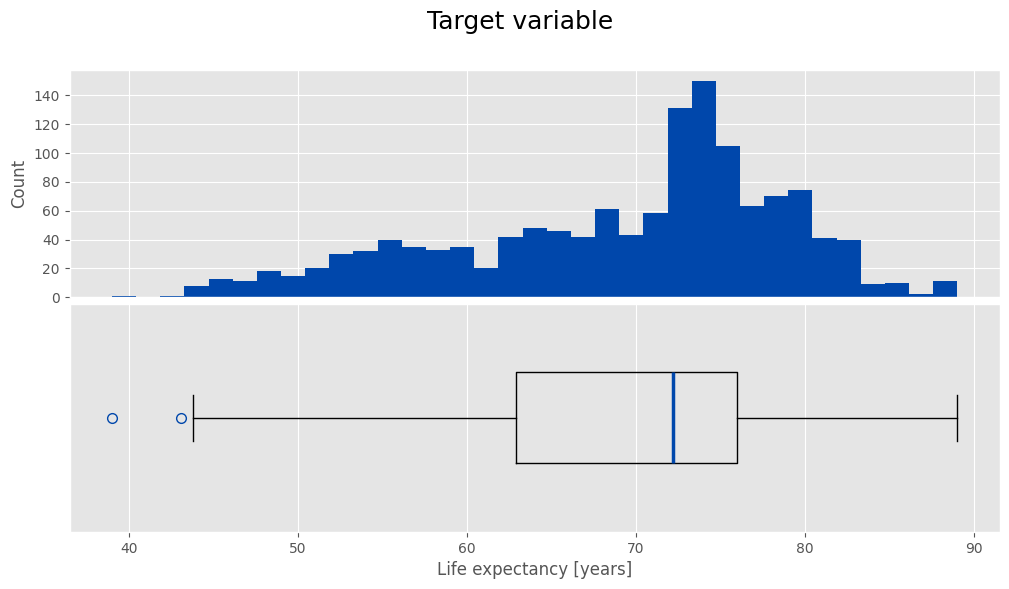
🎯 Target Variable
Here, we examine the target variable and its relationship with other features.
fig, axes = plt.subplots(2, 1, figsize=(12, 6), sharex=True)
fig.subplots_adjust(hspace=0.03)
fig.suptitle('Target variable', size=18)
ax = axes[0]
ax.hist(y_train, bins=35, color=main_color)
ax.set_ylabel('Count')
ax.tick_params(labelbottom=False)
ax.tick_params(axis='x', which='both', length=0)
medianprops = dict(linewidth=2.5, color=main_color)
flierprops = dict(marker='o', markerfacecolor='none', markersize=7, markeredgecolor=main_color)
ax = axes[1]
ax.boxplot(y_train, vert=False, widths=[0.4], showfliers=True, flierprops=flierprops, medianprops=medianprops)
ax.get_yaxis().set_visible(False)
ax.set_xlabel('Life expectancy [years]')
plt.show()
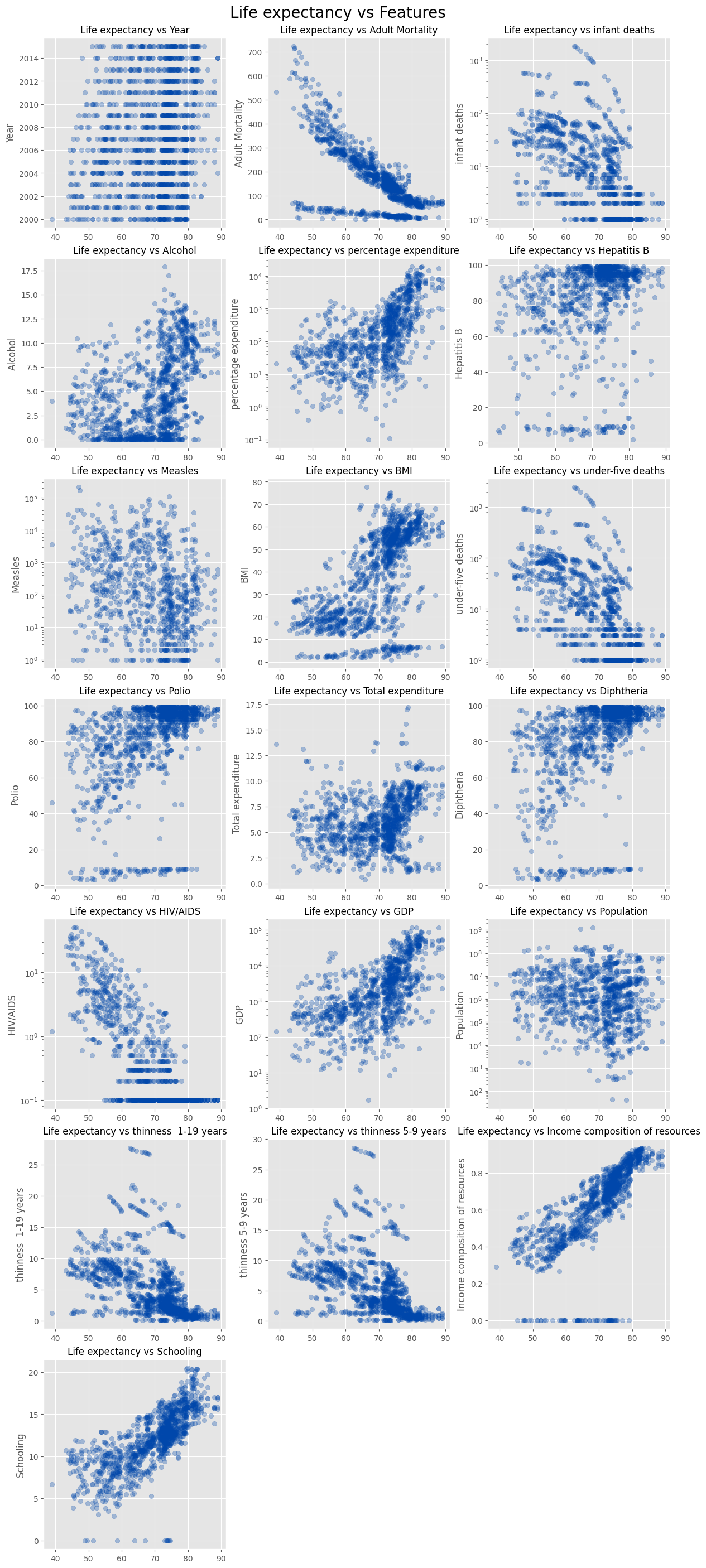
We analyze relationships between features, specifically identifying highly correlated ones for later special handling.
fig, axes = plt.subplots(7, 3, figsize=(12, 28), layout='constrained')
fig.suptitle('Life expectancy vs Features', size=20)
# fig.supxlabel('Life expectancy', size=18)
alpha = 0.3
for col, ax in zip(num_columns, fig.axes):
data = X_train[col]
if col in logscaled_columns:
ax.set_yscale('log')
ax.scatter(y_train, data, alpha=alpha, color=main_color)
else:
ax.scatter(y_train, data, alpha=alpha, color=main_color)
ax.set_ylabel(col)
ax.set_title(f'Life expectancy vs {col}', fontsize=12)
axes[6][1].axis('off')
axes[6][2].axis('off')
plt.show()
colors_r = plt.get_cmap('Blues')(np.linspace(0, 1, 128))
colors_l = colors_r[::-1]
ggcmap_bi = mcolors.LinearSegmentedColormap.from_list('ggplot_like', np.vstack((colors_l, colors_r)))
fig, ax = plt.subplots(figsize=(12, 8), layout='constrained')
fig.suptitle('Correlation matrix', size=20)
corr = X_train[num_columns].corr()
sns.heatmap(corr.round(1), ax=ax, square=True, annot=True, linewidths=0.3,
annot_kws={"size": 8}, cmap=ggcmap_bi, vmin=-1, vmax=1)
ax.tick_params(axis='both', which='both',length=0)
plt.show()
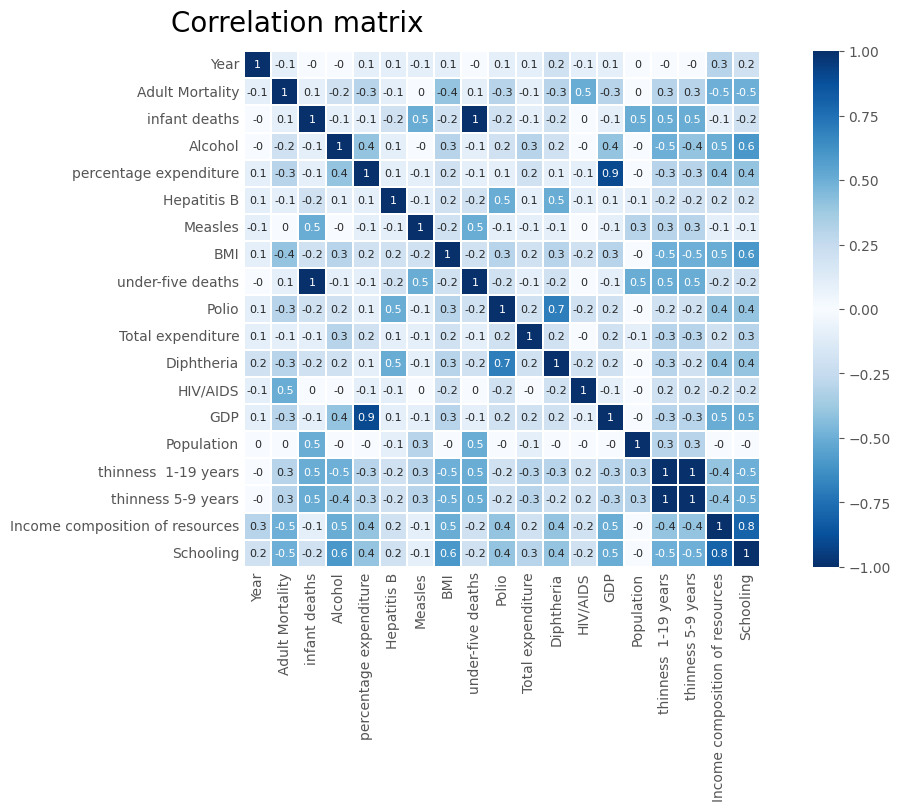
high_corr_columns = ['under-five deaths', 'thinness 5-9 years', 'GDP']
🏭 Feature Transformations
This section prepares the feature preprocessing steps, including handling missing values, logarithmic transformations, and encoding. A new continent feature is also created.
🔧 Transformation Preparation
🗂️ Categorical Features
One-Hot Encoding (OHE) is prepared for categorical features.
X_train[cat_columns].describe().T
| count | unique | top | freq | |
|---|---|---|---|---|
| Status | 1358 | 2 | Developing | 1108 |
| Country | 1358 | 183 | Afghanistan | 13 |
cat_encoder = Pipeline([
('ohe_encoder', OneHotEncoder(handle_unknown='ignore')),
])
🔢 Numerical Features
An imputer is prepared to handle missing values in numerical features.
X_train[num_columns].describe().T
| count | mean | std | min | 25% | 50% | 75% | max | |
|---|---|---|---|---|---|---|---|---|
| Year | 1358.0 | 2.007104e+03 | 4.535324e+00 | 2000.00000 | 2003.000000 | 2.007000e+03 | 2.011000e+03 | 2.015000e+03 |
| Adult Mortality | 1358.0 | 1.660037e+02 | 1.273147e+02 | 1.00000 | 73.000000 | 1.420000e+02 | 2.290000e+02 | 7.230000e+02 |
| infant deaths | 1358.0 | 3.252651e+01 | 1.300906e+02 | 0.00000 | 0.000000 | 3.000000e+00 | 2.300000e+01 | 1.800000e+03 |
| Alcohol | 1278.0 | 4.806667e+00 | 4.091876e+00 | 0.01000 | 0.962500 | 4.055000e+00 | 8.042500e+00 | 1.787000e+01 |
| percentage expenditure | 1358.0 | 8.191974e+02 | 2.179784e+03 | 0.00000 | 7.342322 | 6.750684e+01 | 4.571048e+02 | 1.909905e+04 |
| Hepatitis B | 1098.0 | 8.152823e+01 | 2.407928e+01 | 2.00000 | 77.000000 | 9.200000e+01 | 9.600000e+01 | 9.900000e+01 |
| Measles | 1358.0 | 2.237757e+03 | 1.072481e+04 | 0.00000 | 0.000000 | 1.700000e+01 | 3.517500e+02 | 2.121830e+05 |
| BMI | 1346.0 | 3.818388e+01 | 1.996276e+01 | 1.00000 | 18.925000 | 4.325000e+01 | 5.610000e+01 | 7.760000e+01 |
| under-five deaths | 1358.0 | 4.508689e+01 | 1.772966e+02 | 0.00000 | 0.000000 | 4.000000e+00 | 3.175000e+01 | 2.500000e+03 |
| Polio | 1350.0 | 8.232741e+01 | 2.357581e+01 | 3.00000 | 77.250000 | 9.300000e+01 | 9.700000e+01 | 9.900000e+01 |
| Total expenditure | 1262.0 | 6.037964e+00 | 2.425639e+00 | 0.37000 | 4.370000 | 5.845000e+00 | 7.700000e+00 | 1.720000e+01 |
| Diphtheria | 1350.0 | 8.246296e+01 | 2.356940e+01 | 3.00000 | 78.000000 | 9.250000e+01 | 9.700000e+01 | 9.900000e+01 |
| HIV/AIDS | 1358.0 | 1.904271e+00 | 5.498987e+00 | 0.10000 | 0.100000 | 1.000000e-01 | 8.000000e-01 | 5.060000e+01 |
| GDP | 1170.0 | 7.999548e+03 | 1.527320e+04 | 1.68135 | 453.543922 | 1.705259e+03 | 6.476837e+03 | 1.157616e+05 |
| Population | 1079.0 | 1.234136e+07 | 5.776179e+07 | 41.00000 | 187528.500000 | 1.373513e+06 | 7.417429e+06 | 1.293859e+09 |
| thinness 1-19 years | 1346.0 | 4.997845e+00 | 4.645902e+00 | 0.10000 | 1.600000 | 3.400000e+00 | 7.400000e+00 | 2.770000e+01 |
| thinness 5-9 years | 1346.0 | 5.060550e+00 | 4.749024e+00 | 0.10000 | 1.525000 | 3.400000e+00 | 7.400000e+00 | 2.860000e+01 |
| Income composition of resources | 1295.0 | 6.269668e-01 | 2.166401e-01 | 0.00000 | 0.489000 | 6.810000e-01 | 7.840000e-01 | 9.380000e-01 |
| Schooling | 1295.0 | 1.207097e+01 | 3.396963e+00 | 0.00000 | 10.100000 | 1.240000e+01 | 1.440000e+01 | 2.050000e+01 |
num_encoder = Pipeline([
# ('knn_imputer', KNNImputer(missing_values=np.nan, n_neighbors=5, weights='distance')),
('mean_imputer', SimpleImputer(missing_values=np.nan, strategy='mean')),
])
🪵 Logarithmic Features
Values for features with a logarithmic scale are transformed.
def log_transform(x):
return np.log(x + 1)
log_encoder = Pipeline([
# ('knn_imputer', KNNImputer(missing_values=np.nan, n_neighbors=5, weights='distance')),
('mean_imputer', SimpleImputer(missing_values=np.nan, strategy='mean')),
('log_transformer', FunctionTransformer(log_transform))
])
🌍 Continent Feature
A new feature indicating the continent for each country is created.
def country_to_continent(country):
try:
alpha2 = pcc.country_name_to_country_alpha2(country)
continent = pcc.country_alpha2_to_continent_code(alpha2)
return continent
except KeyError:
return np.nan
class ContinentTransformer(BaseEstimator, TransformerMixin):
def __init__(self):
pass
def fit(self, X=None, y=None):
return self
def transform(self, X, y=None):
X['Country'] = X['Country'].apply(country_to_continent)
return X
continent_encoder = Pipeline([
('continent_transformer', ContinentTransformer()),
('ohe_encoder', OneHotEncoder(handle_unknown='ignore')),
])
🗜️ Applying Transformations
This section sets up imputer and scaler transformations on the training data and creates suitable preprocessors for each model.
🌳 Random Forest Preprocessor
column_transformer = ColumnTransformer([
# ('continent_encoder', continent_encoder, ['Country']),
('categorical_encoder', cat_encoder, cat_columns),
('numerical_encoder', num_encoder, num_columns),
])
preprocessor_random_forest = Pipeline([
('preprocessor', column_transformer),
])
preprocessor_random_forest.fit(X_train)
Pipeline(steps=[('preprocessor',
ColumnTransformer(transformers=[('categorical_encoder',
Pipeline(steps=[('ohe_encoder',
OneHotEncoder(handle_unknown='ignore'))]),
['Status', 'Country']),
('numerical_encoder',
Pipeline(steps=[('mean_imputer',
SimpleImputer())]),
['Year', 'Adult Mortality',
'infant deaths', 'Alcohol',
'percentage expenditure',
'Hepatitis B', 'Measles',
'BMI', 'under-five deaths',
'Polio', 'Total expenditure',
'Diphtheria', 'HIV/AIDS',
'GDP', 'Population',
'thinness 1-19 years',
'thinness 5-9 years',
'Income composition of '
'resources',
'Schooling'])]))])</pre><b>In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. <br />On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.</b></div><div class="sk-container" hidden><div class="sk-item sk-dashed-wrapped"><div class="sk-label-container"><div class="sk-label sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-1" type="checkbox" ><label for="sk-estimator-id-1" class="sk-toggleable__label sk-toggleable__label-arrow">Pipeline</label><div class="sk-toggleable__content"><pre>Pipeline(steps=[('preprocessor',
ColumnTransformer(transformers=[('categorical_encoder',
Pipeline(steps=[('ohe_encoder',
OneHotEncoder(handle_unknown='ignore'))]),
['Status', 'Country']),
('numerical_encoder',
Pipeline(steps=[('mean_imputer',
SimpleImputer())]),
['Year', 'Adult Mortality',
'infant deaths', 'Alcohol',
'percentage expenditure',
'Hepatitis B', 'Measles',
'BMI', 'under-five deaths',
'Polio', 'Total expenditure',
'Diphtheria', 'HIV/AIDS',
'GDP', 'Population',
'thinness 1-19 years',
'thinness 5-9 years',
'Income composition of '
'resources',
'Schooling'])]))])</pre></div></div></div><div class="sk-serial"><div class="sk-item sk-dashed-wrapped"><div class="sk-label-container"><div class="sk-label sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-2" type="checkbox" ><label for="sk-estimator-id-2" class="sk-toggleable__label sk-toggleable__label-arrow">preprocessor: ColumnTransformer</label><div class="sk-toggleable__content"><pre>ColumnTransformer(transformers=[('categorical_encoder',
Pipeline(steps=[('ohe_encoder',
OneHotEncoder(handle_unknown='ignore'))]),
['Status', 'Country']),
('numerical_encoder',
Pipeline(steps=[('mean_imputer',
SimpleImputer())]),
['Year', 'Adult Mortality', 'infant deaths',
'Alcohol', 'percentage expenditure',
'Hepatitis B', 'Measles', 'BMI',
'under-five deaths', 'Polio',
'Total expenditure', 'Diphtheria', 'HIV/AIDS',
'GDP', 'Population', 'thinness 1-19 years',
'thinness 5-9 years',
'Income composition of resources',
'Schooling'])])</pre></div></div></div><div class="sk-parallel"><div class="sk-parallel-item"><div class="sk-item"><div class="sk-label-container"><div class="sk-label sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-3" type="checkbox" ><label for="sk-estimator-id-3" class="sk-toggleable__label sk-toggleable__label-arrow">categorical_encoder</label><div class="sk-toggleable__content"><pre>['Status', 'Country']</pre></div></div></div><div class="sk-serial"><div class="sk-item"><div class="sk-serial"><div class="sk-item"><div class="sk-estimator sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-4" type="checkbox" ><label for="sk-estimator-id-4" class="sk-toggleable__label sk-toggleable__label-arrow">OneHotEncoder</label><div class="sk-toggleable__content"><pre>OneHotEncoder(handle_unknown='ignore')</pre></div></div></div></div></div></div></div></div><div class="sk-parallel-item"><div class="sk-item"><div class="sk-label-container"><div class="sk-label sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-5" type="checkbox" ><label for="sk-estimator-id-5" class="sk-toggleable__label sk-toggleable__label-arrow">numerical_encoder</label><div class="sk-toggleable__content"><pre>['Year', 'Adult Mortality', 'infant deaths', 'Alcohol', 'percentage expenditure', 'Hepatitis B', 'Measles', 'BMI', 'under-five deaths', 'Polio', 'Total expenditure', 'Diphtheria', 'HIV/AIDS', 'GDP', 'Population', 'thinness 1-19 years', 'thinness 5-9 years', 'Income composition of resources', 'Schooling']</pre></div></div></div><div class="sk-serial"><div class="sk-item"><div class="sk-serial"><div class="sk-item"><div class="sk-estimator sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-6" type="checkbox" ><label for="sk-estimator-id-6" class="sk-toggleable__label sk-toggleable__label-arrow">SimpleImputer</label><div class="sk-toggleable__content"><pre>SimpleImputer()</pre></div></div></div></div></div></div></div></div></div></div></div></div></div></div>
⛰️ Ridge Regression Preprocessor
column_transformer = ColumnTransformer([
('continent_encoder', continent_encoder, ['Country']),
('categorical_encoder', cat_encoder, cat_columns),
('numerical_encoder', num_encoder, [col for col in num_columns if col not in [high_corr_columns]]),
('logscaled_encoder', log_encoder, logscaled_columns),
])
preprocessor_ridge = Pipeline([
('preprocessor', column_transformer),
# ('basis_functions', PolynomialFeatures(include_bias=True)),
('scaler', MaxAbsScaler()),
])
preprocessor_ridge.fit(X_train)
Pipeline(steps=[('preprocessor',
ColumnTransformer(transformers=[('continent_encoder',
Pipeline(steps=[('continent_transformer',
ContinentTransformer()),
('ohe_encoder',
OneHotEncoder(handle_unknown='ignore'))]),
['Country']),
('categorical_encoder',
Pipeline(steps=[('ohe_encoder',
OneHotEncoder(handle_unknown='ignore'))]),
['Status', 'Country']),
('numerical_...
'thinness 5-9 years',
'Income composition of '
'resources',
'Schooling']),
('logscaled_encoder',
Pipeline(steps=[('mean_imputer',
SimpleImputer()),
('log_transformer',
FunctionTransformer(func=<function log_transform at 0x7fbbd2fbe480>))]),
['infant deaths',
'percentage expenditure',
'Measles',
'under-five deaths',
'HIV/AIDS', 'GDP',
'Population'])])),
('scaler', MaxAbsScaler())])</pre><b>In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. <br />On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.</b></div><div class="sk-container" hidden><div class="sk-item sk-dashed-wrapped"><div class="sk-label-container"><div class="sk-label sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-7" type="checkbox" ><label for="sk-estimator-id-7" class="sk-toggleable__label sk-toggleable__label-arrow">Pipeline</label><div class="sk-toggleable__content"><pre>Pipeline(steps=[('preprocessor',
ColumnTransformer(transformers=[('continent_encoder',
Pipeline(steps=[('continent_transformer',
ContinentTransformer()),
('ohe_encoder',
OneHotEncoder(handle_unknown='ignore'))]),
['Country']),
('categorical_encoder',
Pipeline(steps=[('ohe_encoder',
OneHotEncoder(handle_unknown='ignore'))]),
['Status', 'Country']),
('numerical_...
'thinness 5-9 years',
'Income composition of '
'resources',
'Schooling']),
('logscaled_encoder',
Pipeline(steps=[('mean_imputer',
SimpleImputer()),
('log_transformer',
FunctionTransformer(func=<function log_transform at 0x7fbbd2fbe480>))]),
['infant deaths',
'percentage expenditure',
'Measles',
'under-five deaths',
'HIV/AIDS', 'GDP',
'Population'])])),
('scaler', MaxAbsScaler())])</pre></div></div></div><div class="sk-serial"><div class="sk-item sk-dashed-wrapped"><div class="sk-label-container"><div class="sk-label sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-8" type="checkbox" ><label for="sk-estimator-id-8" class="sk-toggleable__label sk-toggleable__label-arrow">preprocessor: ColumnTransformer</label><div class="sk-toggleable__content"><pre>ColumnTransformer(transformers=[('continent_encoder',
Pipeline(steps=[('continent_transformer',
ContinentTransformer()),
('ohe_encoder',
OneHotEncoder(handle_unknown='ignore'))]),
['Country']),
('categorical_encoder',
Pipeline(steps=[('ohe_encoder',
OneHotEncoder(handle_unknown='ignore'))]),
['Status', 'Country']),
('numerical_encoder',
Pipeline(steps=[('mean_...
'GDP', 'Population', 'thinness 1-19 years',
'thinness 5-9 years',
'Income composition of resources',
'Schooling']),
('logscaled_encoder',
Pipeline(steps=[('mean_imputer',
SimpleImputer()),
('log_transformer',
FunctionTransformer(func=<function log_transform at 0x7fbbd2fbe480>))]),
['infant deaths', 'percentage expenditure',
'Measles', 'under-five deaths', 'HIV/AIDS',
'GDP', 'Population'])])</pre></div></div></div><div class="sk-parallel"><div class="sk-parallel-item"><div class="sk-item"><div class="sk-label-container"><div class="sk-label sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-9" type="checkbox" ><label for="sk-estimator-id-9" class="sk-toggleable__label sk-toggleable__label-arrow">continent_encoder</label><div class="sk-toggleable__content"><pre>['Country']</pre></div></div></div><div class="sk-serial"><div class="sk-item"><div class="sk-serial"><div class="sk-item"><div class="sk-estimator sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-10" type="checkbox" ><label for="sk-estimator-id-10" class="sk-toggleable__label sk-toggleable__label-arrow">ContinentTransformer</label><div class="sk-toggleable__content"><pre>ContinentTransformer()</pre></div></div></div><div class="sk-item"><div class="sk-estimator sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-11" type="checkbox" ><label for="sk-estimator-id-11" class="sk-toggleable__label sk-toggleable__label-arrow">OneHotEncoder</label><div class="sk-toggleable__content"><pre>OneHotEncoder(handle_unknown='ignore')</pre></div></div></div></div></div></div></div></div><div class="sk-parallel-item"><div class="sk-item"><div class="sk-label-container"><div class="sk-label sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-12" type="checkbox" ><label for="sk-estimator-id-12" class="sk-toggleable__label sk-toggleable__label-arrow">categorical_encoder</label><div class="sk-toggleable__content"><pre>['Status', 'Country']</pre></div></div></div><div class="sk-serial"><div class="sk-item"><div class="sk-serial"><div class="sk-item"><div class="sk-estimator sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-13" type="checkbox" ><label for="sk-estimator-id-13" class="sk-toggleable__label sk-toggleable__label-arrow">OneHotEncoder</label><div class="sk-toggleable__content"><pre>OneHotEncoder(handle_unknown='ignore')</pre></div></div></div></div></div></div></div></div><div class="sk-parallel-item"><div class="sk-item"><div class="sk-label-container"><div class="sk-label sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-14" type="checkbox" ><label for="sk-estimator-id-14" class="sk-toggleable__label sk-toggleable__label-arrow">numerical_encoder</label><div class="sk-toggleable__content"><pre>['Year', 'Adult Mortality', 'infant deaths', 'Alcohol', 'percentage expenditure', 'Hepatitis B', 'Measles', 'BMI', 'under-five deaths', 'Polio', 'Total expenditure', 'Diphtheria', 'HIV/AIDS', 'GDP', 'Population', 'thinness 1-19 years', 'thinness 5-9 years', 'Income composition of resources', 'Schooling']</pre></div></div></div><div class="sk-serial"><div class="sk-item"><div class="sk-serial"><div class="sk-item"><div class="sk-estimator sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-15" type="checkbox" ><label for="sk-estimator-id-15" class="sk-toggleable__label sk-toggleable__label-arrow">SimpleImputer</label><div class="sk-toggleable__content"><pre>SimpleImputer()</pre></div></div></div></div></div></div></div></div><div class="sk-parallel-item"><div class="sk-item"><div class="sk-label-container"><div class="sk-label sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-16" type="checkbox" ><label for="sk-estimator-id-16" class="sk-toggleable__label sk-toggleable__label-arrow">logscaled_encoder</label><div class="sk-toggleable__content"><pre>['infant deaths', 'percentage expenditure', 'Measles', 'under-five deaths', 'HIV/AIDS', 'GDP', 'Population']</pre></div></div></div><div class="sk-serial"><div class="sk-item"><div class="sk-serial"><div class="sk-item"><div class="sk-estimator sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-17" type="checkbox" ><label for="sk-estimator-id-17" class="sk-toggleable__label sk-toggleable__label-arrow">SimpleImputer</label><div class="sk-toggleable__content"><pre>SimpleImputer()</pre></div></div></div><div class="sk-item"><div class="sk-estimator sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-18" type="checkbox" ><label for="sk-estimator-id-18" class="sk-toggleable__label sk-toggleable__label-arrow">FunctionTransformer</label><div class="sk-toggleable__content"><pre>FunctionTransformer(func=<function log_transform at 0x7fbbd2fbe480>)</pre></div></div></div></div></div></div></div></div></div></div><div class="sk-item"><div class="sk-estimator sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-19" type="checkbox" ><label for="sk-estimator-id-19" class="sk-toggleable__label sk-toggleable__label-arrow">MaxAbsScaler</label><div class="sk-toggleable__content"><pre>MaxAbsScaler()</pre></div></div></div></div></div></div></div>
📐 k-NN Preprocessor
column_transformer = ColumnTransformer([
('continent_encoder', continent_encoder, ['Country']),
('categorical_encoder', cat_encoder, ['Status']),
('numerical_encoder', num_encoder, num_columns),
])
preprocessor_knn = Pipeline([
('preprocessor', column_transformer),
('scaler', MaxAbsScaler()),
])
preprocessor_knn.fit(X_train)
Pipeline(steps=[('preprocessor',
ColumnTransformer(transformers=[('continent_encoder',
Pipeline(steps=[('continent_transformer',
ContinentTransformer()),
('ohe_encoder',
OneHotEncoder(handle_unknown='ignore'))]),
['Country']),
('categorical_encoder',
Pipeline(steps=[('ohe_encoder',
OneHotEncoder(handle_unknown='ignore'))]),
['Status']),
('numerical_encoder',
P...steps=[('mean_imputer',
SimpleImputer())]),
['Year', 'Adult Mortality',
'infant deaths', 'Alcohol',
'percentage expenditure',
'Hepatitis B', 'Measles',
'BMI', 'under-five deaths',
'Polio', 'Total expenditure',
'Diphtheria', 'HIV/AIDS',
'GDP', 'Population',
'thinness 1-19 years',
'thinness 5-9 years',
'Income composition of '
'resources',
'Schooling'])])),
('scaler', MaxAbsScaler())])</pre><b>In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. <br />On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.</b></div><div class="sk-container" hidden><div class="sk-item sk-dashed-wrapped"><div class="sk-label-container"><div class="sk-label sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-20" type="checkbox" ><label for="sk-estimator-id-20" class="sk-toggleable__label sk-toggleable__label-arrow">Pipeline</label><div class="sk-toggleable__content"><pre>Pipeline(steps=[('preprocessor',
ColumnTransformer(transformers=[('continent_encoder',
Pipeline(steps=[('continent_transformer',
ContinentTransformer()),
('ohe_encoder',
OneHotEncoder(handle_unknown='ignore'))]),
['Country']),
('categorical_encoder',
Pipeline(steps=[('ohe_encoder',
OneHotEncoder(handle_unknown='ignore'))]),
['Status']),
('numerical_encoder',
P...steps=[('mean_imputer',
SimpleImputer())]),
['Year', 'Adult Mortality',
'infant deaths', 'Alcohol',
'percentage expenditure',
'Hepatitis B', 'Measles',
'BMI', 'under-five deaths',
'Polio', 'Total expenditure',
'Diphtheria', 'HIV/AIDS',
'GDP', 'Population',
'thinness 1-19 years',
'thinness 5-9 years',
'Income composition of '
'resources',
'Schooling'])])),
('scaler', MaxAbsScaler())])</pre></div></div></div><div class="sk-serial"><div class="sk-item sk-dashed-wrapped"><div class="sk-label-container"><div class="sk-label sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-21" type="checkbox" ><label for="sk-estimator-id-21" class="sk-toggleable__label sk-toggleable__label-arrow">preprocessor: ColumnTransformer</label><div class="sk-toggleable__content"><pre>ColumnTransformer(transformers=[('continent_encoder',
Pipeline(steps=[('continent_transformer',
ContinentTransformer()),
('ohe_encoder',
OneHotEncoder(handle_unknown='ignore'))]),
['Country']),
('categorical_encoder',
Pipeline(steps=[('ohe_encoder',
OneHotEncoder(handle_unknown='ignore'))]),
['Status']),
('numerical_encoder',
Pipeline(steps=[('mean_imputer',
SimpleImputer())]),
['Year', 'Adult Mortality', 'infant deaths',
'Alcohol', 'percentage expenditure',
'Hepatitis B', 'Measles', 'BMI',
'under-five deaths', 'Polio',
'Total expenditure', 'Diphtheria', 'HIV/AIDS',
'GDP', 'Population', 'thinness 1-19 years',
'thinness 5-9 years',
'Income composition of resources',
'Schooling'])])</pre></div></div></div><div class="sk-parallel"><div class="sk-parallel-item"><div class="sk-item"><div class="sk-label-container"><div class="sk-label sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-22" type="checkbox" ><label for="sk-estimator-id-22" class="sk-toggleable__label sk-toggleable__label-arrow">continent_encoder</label><div class="sk-toggleable__content"><pre>['Country']</pre></div></div></div><div class="sk-serial"><div class="sk-item"><div class="sk-serial"><div class="sk-item"><div class="sk-estimator sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-23" type="checkbox" ><label for="sk-estimator-id-23" class="sk-toggleable__label sk-toggleable__label-arrow">ContinentTransformer</label><div class="sk-toggleable__content"><pre>ContinentTransformer()</pre></div></div></div><div class="sk-item"><div class="sk-estimator sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-24" type="checkbox" ><label for="sk-estimator-id-24" class="sk-toggleable__label sk-toggleable__label-arrow">OneHotEncoder</label><div class="sk-toggleable__content"><pre>OneHotEncoder(handle_unknown='ignore')</pre></div></div></div></div></div></div></div></div><div class="sk-parallel-item"><div class="sk-item"><div class="sk-label-container"><div class="sk-label sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-25" type="checkbox" ><label for="sk-estimator-id-25" class="sk-toggleable__label sk-toggleable__label-arrow">categorical_encoder</label><div class="sk-toggleable__content"><pre>['Status']</pre></div></div></div><div class="sk-serial"><div class="sk-item"><div class="sk-serial"><div class="sk-item"><div class="sk-estimator sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-26" type="checkbox" ><label for="sk-estimator-id-26" class="sk-toggleable__label sk-toggleable__label-arrow">OneHotEncoder</label><div class="sk-toggleable__content"><pre>OneHotEncoder(handle_unknown='ignore')</pre></div></div></div></div></div></div></div></div><div class="sk-parallel-item"><div class="sk-item"><div class="sk-label-container"><div class="sk-label sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-27" type="checkbox" ><label for="sk-estimator-id-27" class="sk-toggleable__label sk-toggleable__label-arrow">numerical_encoder</label><div class="sk-toggleable__content"><pre>['Year', 'Adult Mortality', 'infant deaths', 'Alcohol', 'percentage expenditure', 'Hepatitis B', 'Measles', 'BMI', 'under-five deaths', 'Polio', 'Total expenditure', 'Diphtheria', 'HIV/AIDS', 'GDP', 'Population', 'thinness 1-19 years', 'thinness 5-9 years', 'Income composition of resources', 'Schooling']</pre></div></div></div><div class="sk-serial"><div class="sk-item"><div class="sk-serial"><div class="sk-item"><div class="sk-estimator sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-28" type="checkbox" ><label for="sk-estimator-id-28" class="sk-toggleable__label sk-toggleable__label-arrow">SimpleImputer</label><div class="sk-toggleable__content"><pre>SimpleImputer()</pre></div></div></div></div></div></div></div></div></div></div><div class="sk-item"><div class="sk-estimator sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-29" type="checkbox" ><label for="sk-estimator-id-29" class="sk-toggleable__label sk-toggleable__label-arrow">MaxAbsScaler</label><div class="sk-toggleable__content"><pre>MaxAbsScaler()</pre></div></div></div></div></div></div></div>
🌳 Random Forest
🏗️ Model Implementation
class CustomRandomForest(RegressorMixin):
"""
Custom Random Forest Regressor.
This model utilizes DecisionTreeRegressor from sklearn as its base estimators.
"""
def __init__(self, n_estimators, max_samples_fraction, max_depth, **kwargs):
"""
Model constructor.
Key hyperparameters:
n_estimators: Number of decision tree sub-models.
max_samples_fraction: The fraction of samples to bootstrap for each sub-model (0 to 1).
max_depth: Maximum depth of each decision tree sub-model.
kwargs: (Optional) Additional hyperparameters for the DecisionTreeRegressor sub-models.
"""
self.n_estimators = n_estimators
self.max_samples_fraction = max_samples_fraction
self.max_depth = max_depth
self.decision_tree_kwargs = kwargs
def fit(self, X, y):
"""
Trains the model. Training data is provided in X and y.
Sub-models are trained using bootstrapping, with sample size determined by max_samples_fraction.
"""
self.estimators = []
n_samples = int(X.shape[0] * self.max_samples_fraction)
for _ in range(self.n_estimators):
X_sample, y_sample = resample(X, y, replace=True, n_samples=n_samples)
tree = DecisionTreeRegressor(splitter='random', max_depth=self.max_depth, **self.decision_tree_kwargs)
tree.fit(X_sample, y_sample)
self.estimators.append(tree)
def predict(self, X):
"""
Predicts y for the given data points in X.
"""
estimations = np.zeros((self.n_estimators, X.shape[0]))
for i, estimator in enumerate(self.estimators):
estimations[i] = estimator.predict(X)
y_predicted = estimations.mean(axis=0)
return y_predicted
👍 Model Suitability
The Random Forest model is well-suited for this dataset due to its robustness.
- It handles outliers and various feature types effectively.
- The random feature selection in its implementation leads to diverse trees, promoting good generalization.
- Unlike single decision trees, it’s less sensitive to data changes and generally yields strong results.
- It requires minimal preprocessing and has a lower risk of overfitting during training.
- While training time is longer (though parallelizable), its interpretability is lower compared to a single tree.
⏱️ Hyperparameters for Tuning
param_grid = ParameterGrid({
'n_estimators': range(20, 300, 20),
})
✨ Best Model Selection
X_train_np = preprocessor_random_forest.transform(X_train)
X_val_np = preprocessor_random_forest.transform(X_val)
log_rf = pd.DataFrame(columns=['n_estimators', 'train_rmse', 'val_rmse'])
estimators_rf = []
for params in param_grid:
reg = CustomRandomForest(max_depth=None, max_samples_fraction=1, **params)
reg.fit(X_train_np, y_train)
train_rmse = mse(y_train, reg.predict(X_train_np), squared=False)
val_rmse = mse(y_val, reg.predict(X_val_np), squared=False)
log_rf.loc[len(log_rf.index)] = [params['n_estimators'], train_rmse, val_rmse]
estimators_rf.append(reg)
log_rf.sort_index(inplace=True)
fig, axes = plt.subplots(2, 1, figsize=(12, 5), layout='constrained', sharex=True)
fig.suptitle('Random forest learning curve', size=20)
fig.supylabel('RMSE', size=16)
fig.supxlabel('Number of estimators', size=16)
ax = axes[0]
ax.plot(log_rf['n_estimators'], log_rf['train_rmse'], label='train', color=main_color, linewidth=2.5)
ax.legend()
ax = axes[1]
ax.plot(log_rf['n_estimators'], log_rf['val_rmse'], label='validation', color='green', linewidth=2.5)
ax.legend()
plt.show()
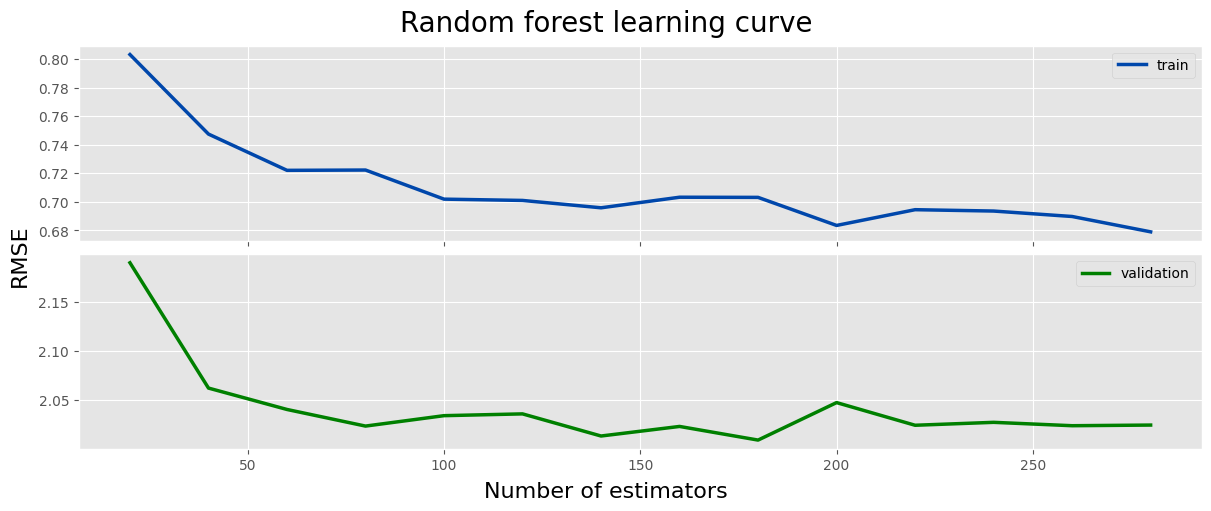
log_rf = log_rf.sort_values('val_rmse')
log_rf.head(5)
n_estimators train_rmse val_rmse 8 180.0 0.703142 2.009168 6 140.0 0.695845 2.013311 7 160.0 0.703251 2.023062 3 80.0 0.722281 2.023428 12 260.0 0.689763 2.023804
best_rf = estimators_rf[log_rf.sort_values('val_rmse').index[0]]
📈 Best Model Evaluation
X_val_np = preprocessor_random_forest.transform(X_val)
rf_eval = pd.DataFrame([
mse(y_val, best_rf.predict(X_val_np), squared=False),
mae(y_val, best_rf.predict(X_val_np)),
], index=['RMSE', 'MAE'], columns=['random forest'])
rf_eval
alpha train_rmse val_rmse 8 0.000808 1.592143 2.146361 9 0.000909 1.593479 2.146424 7 0.000707 1.590830 2.146496 10 0.001010 1.594850 2.146761 11 0.001111 1.596189 2.146996
best_ridge = estimators_ridge[log_ridge.sort_values('val_rmse').index[0]]
📈 Best Model Evaluation
X_val_np = preprocessor_ridge.transform(X_val)
ridge_eval = pd.DataFrame([
mse(y_val, best_ridge.predict(X_val_np), squared=False),
mae(y_val, best_ridge.predict(X_val_np)),
], index=['RMSE', 'MAE'], columns=['ridge'])
ridge_eval
ridge RMSE 2.146361 MAE 1.222629
📐 k-NN regression
👍 Model Suitability
k-Nearest Neighbors (k-NN) is a suitable method for this task:
- k-NN is a non-parametric model that doesn’t require explicit training; it simply stores the training data.
- While prediction can be computationally intensive for large datasets, it’s not an issue here due to the relatively small training data size.
- Given that the data is normalized and one-hot encoded, and the data dimensionality is relatively low (avoiding the curse of dimensionality), k-NN can be an effective approach.
⏱️ Hyperparameters for Tuning
param_grid = ParameterGrid({
'n_neighbors': range(1, 20),
'weights': ['uniform', 'distance'],
})
✨ Best Model Selection
X_train_np = preprocessor_knn.transform(X_train)
X_val_np = preprocessor_knn.transform(X_val)
log_knn = pd.DataFrame(columns=['n_neighbors', 'weights', 'train_rmse', 'val_rmse'])
estimators_knn = []
for params in param_grid:
reg = KNeighborsRegressor(n_neighbors=params['n_neighbors'], weights=params['weights'])
reg.fit(X_train_np, y_train)
train_rmse = mse(y_train, reg.predict(X_train_np), squared=False)
val_rmse = mse(y_val, reg.predict(X_val_np), squared=False)
log_knn.loc[len(log_knn.index)] = [params['n_neighbors'], params['weights'], train_rmse, val_rmse]
estimators_knn.append(reg)
log_knn.sort_index(inplace=True)
df = log_knn[log_knn['weights'] == 'distance']
fig, axes = plt.subplots(2, 1, figsize=(12, 5), layout='constrained', sharex=True)
fig.suptitle('KNN learning curve (distance)', size=20)
fig.supylabel('RMSE', size=16)
fig.supxlabel('Number of neighbors', size=16)
ax = axes[0]
ax.plot(df['n_neighbors'], df['train_rmse'], label='train', color=main_color, linewidth=2.5)
ax.legend(loc='lower right')
ax = axes[1]
ax.plot(df['n_neighbors'], df['val_rmse'], label='validation', color='chocolate', linewidth=2.5)
ax.legend(loc='lower right')
ax.xaxis.set_major_locator(MaxNLocator(integer=True))
plt.show()
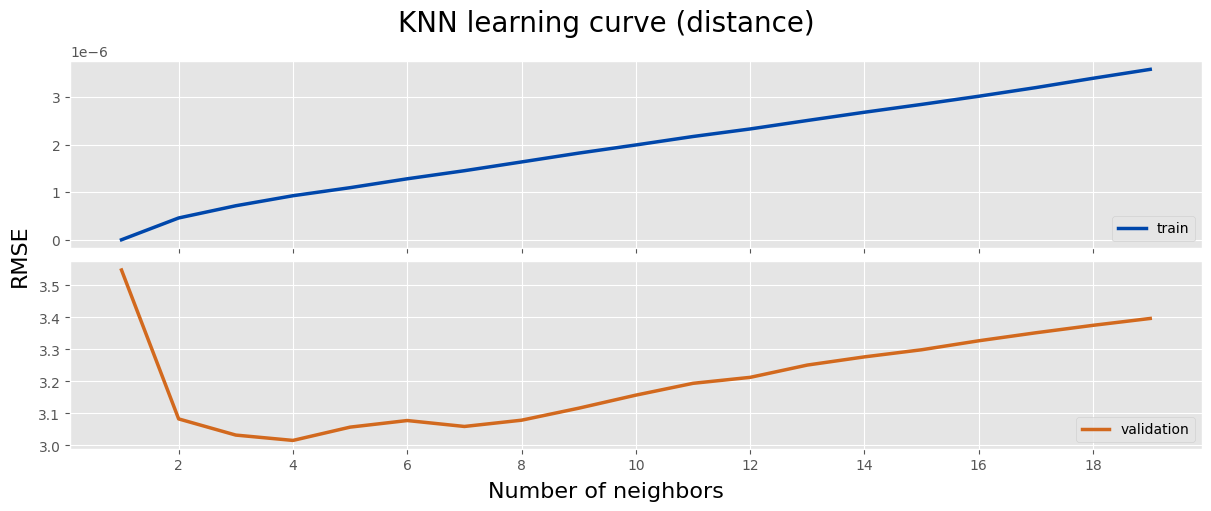
log_knn = log_knn.sort_values('val_rmse')
log_knn.head(5)
n_neighbors weights train_rmse val_rmse 7 4 distance 9.274417e-07 3.015674 5 3 distance 7.163777e-07 3.032370 9 5 distance 1.097045e-06 3.057202 13 7 distance 1.454370e-06 3.059293 11 6 distance 1.284174e-06 3.077836
best_knn = estimators_knn[log_knn.sort_values('val_rmse').index[0]]
📈 Best Model Evaluation
X_val_np = preprocessor_knn.transform(X_val)
knn_eval = pd.DataFrame([
mse(y_val, best_knn.predict(X_val_np), squared=False),
mae(y_val, best_knn.predict(X_val_np)),
], index=['RMSE', 'MAE'], columns=['knn'])
knn_eval
knn RMSE 3.015674 MAE 1.878656
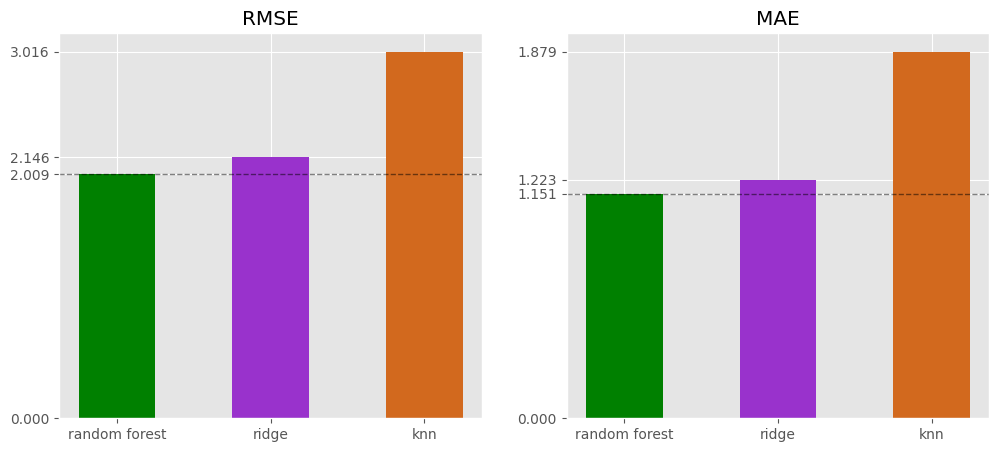
⚖️ Model Comparison
This section compares the best models from each family, selects the model with the lowest RMSE, retrains it on the full training data (training + validation), and estimates the RMSE on new data using the test set.
eval = pd.concat([rf_eval.T, ridge_eval.T, knn_eval.T])
eval
RMSE MAE random forest 2.009168 1.151335 ridge 2.146361 1.222629 knn 3.015674 1.878656
fig, axes = plt.subplots(1, 2, figsize=(12, 5))
ax = axes[0]
for metric, ax in zip(['RMSE', 'MAE'], fig.axes):
ax.set_title(metric)
bars = ax.bar(eval.index, eval.loc[:, metric], color=['green', 'darkorchid', 'chocolate'], width=0.5)
ax.axhline(bars[0].get_height(), color = 'black', linestyle = '--', alpha=0.5, linewidth=1)
ax.set_yticks([0, *[b.get_height() for b in bars]])
🏆 Final Model
Based on the validation data, the Random Forest model demonstrates the best performance.
The chosen model will now be retrained on the combined training and validation data. Its objective RMSE on new, unseen data will then be measured using the test set.
X_train_val_np = preprocessor_random_forest.fit_transform(X_train_val)
best_rf.fit(X_train_val_np, y_train_val)
X_test_np = preprocessor_random_forest.transform(X_test)
print(mse(y_test, best_rf.predict(X_test_np), squared=False))
1.7480988972377174
We expect an approximate RMSE of 1.75 on new data.
🎯 Evaluating evaluation.csv
eval_data = pd.read_csv('evaluation.csv')
eval_data_np = preprocessor_random_forest.transform(eval_data)
eval_data['Life expectancy'] = best_rf.predict(eval_data_np)
eval_data.to_csv('results.csv', columns=['Country', 'Year', 'Life expectancy'], header=True, index=False)